Abstract
Despite more than 50 years of vaccination, Bordetella pertussis has remained endemic in The Netherlands, causing epidemic outbreaks every 3 to 5 years. Strain variation may play a role in the persistence of B. pertussis and was studied by sequencing 15 genes coding for surface proteins, including genes for all five components of acellular pertussis vaccines: pertussis toxin (Ptx), pertactin (Prn), filamentous hemagglutinin, and fimbriae (Fim2 and Fim3). A low level of allelic variation was observed, confirming a recent evolutionary origin of B. pertussis. In modern isolates, polymorphism was observed only in prn, ptxS1, ptxS3, and tcfA. Polymorphism in ptxS1, ptxS3, and tcfA was used to categorize isolates in multilocus sequence types (MLSTs). Analysis of Dutch isolates from 1949 to 1999 revealed five MLSTs, which showed a highly dynamic temporal behavior. We observed significant changes in the MLSTs after the introduction of pertussis vaccination in The Netherlands. Epidemic years were found to be associated with the expansion of MLST-4 or MLST-5. MLST-5 showed a remarkable expansion from 10% in 1997 to 80% in 1999. The MLST analysis was extended to a number of widely separated geographic regions: Finland, Italy, Japan, and the United States. MLST-4 and MLST-5 were found to dominate in Italy and the United States. In Finland and Japan, MLST-3 and MLST-2, respectively, were predominant. Thus, although each region showed distinctive MLST frequencies, in three of the five regions MLST-4 and MLST-5 were predominant. These types may represent newly emerged, successful clones. The identification of highly successful clones may shed light on the question of how B. pertussis is able to maintain itself in vaccinated populations.
Despite more than 50 years of vaccination, pertussis has remained an endemic disease, with epidemic outbreaks occurring every 3 to 5 years. In some vaccinated populations (e.g., in Australia, Canada, The Netherlands, and the United States) the incidence of pertussis is increasing (1, 3, 7-9, 17). The resilience of Bordetella pertussis to vaccination may be due to its highly infectious nature, its ability to hamper the host immune response, the shortcomings of pertussis vaccines, and strain variation. Our studies on strain variation have led us to propose that the resurgence of pertussis may be due, in part, to the adaptation of the B. pertussis population to vaccination (26). The aim of the present study was twofold. First, we sought to extend these studies and to find markers suitable for the analysis of B. pertussis populations. Second, we sought to determine the extent of polymorphism in immunologically relevant proteins, in particular those incorporated in acellular vaccines (ACVs).
Recently, multilocus sequence typing (MLST) has been introduced as a new approach for studying the molecular epidemiology of bacterial pathogens (11, 30). MLST is based on the well-tested principles of multilocus enzyme electrophoresis, but it assigns alleles at each site directly by nucleotide sequencing rather than indirectly from electrophoretic mobilities of their gene products in starch gels. An important advantage of MLST over the other typing methods, such as restriction fragment length polymorphism (RFLP), randomly amplified polymorphic DNA, and pulsed-field gel electrophoresis, is that the sequence data are truly comparable between laboratories (23). Further, MLST is more amenable to quantitative analyses, allowing the establishment of quantitative genetic relationships between isolates. Finally, MLST has proven to be especially suitable for studying longer-term and global epidemiology (10, 12, 19).
B. pertussis is a very homogeneous species and, consequently, finding polymorphic sites for MLST has proved to be difficult. Multilocus enzyme electrophoresis revealed only four electrophoretic types (27, 34). Consistent with this, we found very little polymorphism in B. pertussis genes coding for housekeeping genes (unpublished data). In view of the restricted polymorphism found in B. pertussis, we attempted to increase the likelihood of identifying allelic variation by analysis of genes for surface proteins, which may be subject to more intensive selective pressure. In previous work we showed that the S1 subunit of pertussis toxin (PtxS1) and pertactin (Prn) were polymorphic (4, 24-26). PtxS1 and Prn comprise two of the five B. pertussis proteins of ACVs. In this work, we investigated polymorphism in the remaining B. pertussis proteins, which are included in ACVs: the pertussis toxin subunits PtxS2, PtxS3, PtxS4, and PtxS5; the filamentous hemagglutinin (FHA); and serotype 2 and 3 fimbriae (Fim2 and Fim3). Further, we analyzed variation in a number of other surface-associated proteins, some of which have been shown to be important for immunity: two integral outer membrane proteins, OmpP and OmpQ (2, 22), tracheal colonization factor (TcfA) (14), Bordetella resistance to killing protein (BrkA) (29), Vag8 (vir-activated gene 8) (13), and Bordetella intermediate-phase protein (BipA) (32). The polymorphic genes were used to characterize strains from widely separated geographic areas, providing a global perspective. We identified MLSTs associated with epidemics and with strains that have spread globally.
MATERIALS AND METHODS
Strains.
B. pertussis strains were collected from 1949 to 1999. In total, 196 strains were investigated: 128 from The Netherlands, 27 from Finland, 10 from Italy, 13 from Japan, and 18 from the United States. A table, detailing the origin of the strains, is available from the authors. Strains were grown on Bordet-Gengou agar supplemented with 1% glycerol and 4% sheep blood at 35°C for 3 days.
DNA sequencing.
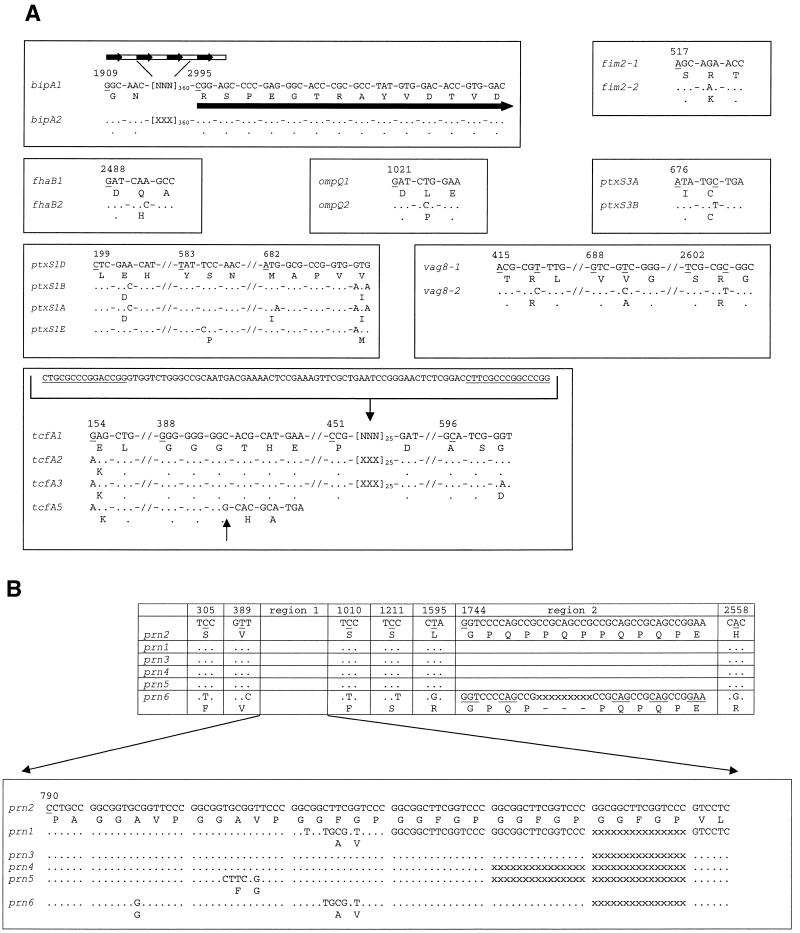
PCR amplification of chromosomal DNA was performed by adding 1 μl of DNA to 19 μl of buffer comprising 50% HotstarTaq Master mix (Qiagen), 1 μM concentrations of each primer, and 5 or 10% dimethyl sulfoxide. Then, 5% dimethyl sulfoxide was added to the PCR mix for amplification of fim2, fim3, fhaB, ompP, ompQ, ptxS1 to ptxS5, and tcfA. For bipA, brkA, prn, and vag8, 10% dimethyl sulfoxide was used. Amplification of genes was performed in a Hybaid Omnigene incubator by using a specific program for each gene. The PCR fragments were purified with a QiaQuick PCR purification kit (Qiagen) and sequenced with the primers used for amplification and internal primers. The primer sequences and PCR programs can be obtained from the authors. Sequence reactions were performed with an ABI Prism Big Dye terminator reaction kit, and the reactions were analyzed with a model 377 or 3700 ABI DNA Sequencer (Perkin-Elmer Applied Biosystems). For bipA, brkA, fim2, fim3, ompP, ompQ, ptxS1, ptxS2, ptxS3, ptxS4, ptxS5, and vag8, the complete open reading frame was sequenced. The fhaB gene from 13 strains was sequenced between bases 1 and 6612. This region codes for the secreted, processed molecule incorporated in ACVs. Subsequently, sequencing of fhaB was confined to the region comprising bases 2250 to 2750, which contained the single polymorphic site identified. The tcfA gene from five strains was sequenced completely. Two polymorphic regions were identified between bases 1 and 945. A primary region, comprised of bases 395 to 945, was sequenced for all strains. The secondary region, comprised of bases 1 to 395, appeared to be linked to the primary region and was noninformative. Consequently, the secondary region was sequenced for 20% of the Dutch strains but for all strains outside The Netherlands. Previously, two polymorphic regions (region 1 and 2) were identified in prn, and essentially all polymorphism was found to be restricted to region 1 (26). Region 1 was sequenced for all strains, whereas region 2 was sequenced for 20% of the strains.
RESULTS
Polymorphism in B. pertussis genes coding for surface proteins.
We investigated polymorphism in 15 genes, coding for surface proteins (Table 1). Each gene was sequenced completely for the two strains used for the WCV and three recent Dutch clinical isolates. Assuming that vaccination had selected for strains that are antigenically distinct from the vaccine strains, this approach was expected to increase the likelihood of finding polymorphism. Further, we included strain 18323 in our analyses, which represents a distinct branch in the phylogenetic tree of B. pertussis (27, 34). No polymorphism was found in brkA, fim3, ompP, ptxS2, ptxS4, and ptxS5. Polymorphism was observed in bipA, fhaB, fim2, ompQ, ptxS1, ptxS3, prn, tcfA, and vag8 (Fig. 1). Data from two genes, ptxS1 and prn, were mainly from previous studies (4, 24-26). However, here we extended previous data by analyzing Japanese isolates and Dutch isolates from 1997 to 1999. In all, 22 point mutations were observed in the polymorphic genes, 15 of which were nonsilent. The silent mutations were found in ptxS3 (n = 1), prn (n = 4), and vag8 (n = 2), whereas the nonsilent mutations were observed in fim2 (n = 1), fhaB (n = 1), ompQ (n = 1), ptxS1 (n = 4), prn (n = 5), tcfA (n = 2), and vag8 (n = 1). In prn and bipA, a variable number of repeats was found (Fig. 1). Polymorphism in bipA, fim2, fhaB, ompQ, and vag8 was restricted. Two alleles were found for each of these genes, resulting from polymorphism in one (bipA, fim2, fhaB, and ompQ) or three (vag8) bases (Fig. 1). Further, different alleles for these genes were only observed to coexist in the prevaccination era in The Netherlands (fim2 and fhaB), or variation was only observed relative to the atypical strain 18323 (bipA, ompQ, and vag8). Strain 18323 harbored the following alleles bipA2, fim2-1, fhaB2, ompQ1, prn6, ptxS1E, ptxS3A, tcfA1, and vag8-1, showing divergence with Dutch clinical isolates in all polymorphic sites, except for fim2 and ptxS3 (Fig. 1). In the period from 1990 to 1999, only one allele for bipA (bipA1), fim2 (fim2-1), fhaB (fhaB1), ompQ (ompQ2), and vag8 (vag8-2) was found in the Dutch B. pertussis population.
TABLE 1.
B. pertussis genes analyzed for polymorphism
| Gene | Allele | No. of strains analyzed | % Frequency (n) | GenBank accession no. |
|---|---|---|---|---|
| ptxS1a | 196 | M13223 | ||
| ptxS1A | 79 (154) | AJ245366 | ||
| ptxS1B | 17 (33) | AJ245367 | ||
| ptxS1D | 4 (7) | AJ245368 | ||
| ptxS1E | 1 (2) | AJ006151 | ||
| ptxS2a | 21 | M13223 | ||
| ptxS3a | 196 | |||
| ptxS3A | 78 (153) | M13223 | ||
| ptxS3B | 22 (43) | AJ420987 | ||
| ptxS4a | 22 | M13223 | ||
| ptxS5a | 22 | M13223 | ||
| prnb | 196 | |||
| prn1 | 34 (67) | AJ011091 | ||
| prn2 | 41 (80) | AJ011092 | ||
| prn3 | 22 (43) | AJ011093 | ||
| prn4 | 0.5 (1) | AJ011015 | ||
| prn5 | 1.5 (3) | AJ011016 | ||
| prn6 | 1 (2) | AJ132095 | ||
| fim2a | 19 | |||
| fim2-1 | 84 (16) | Y00527 | ||
| fim2-2 | 16 (3) | AJ420988 | ||
| fim3a | 12 | X51543 | ||
| brkAa | 5 | U12276 | ||
| fhaBc | 22 | |||
| fhaB-1 | 86 (19) | M60351 | ||
| J04531 | ||||
| X52156 | ||||
| fhaB-2 | 14 (3) | AJ420989 | ||
| ompPa | 18 | X58488 | ||
| ompQa | 6 | |||
| ompQ1 | 17 (1) | U16266 | ||
| ompQ2 | 83 (5) | AJ420990 | ||
| tcfAd | 196 | |||
| tcfA1 | 1 (2) | U16754 | ||
| tcfA2 | 80 (157) | AJ009785 | ||
| tcfA3 | 18 (36) | AJ420991 | ||
| tcfA5 | 0.5 (1) | AJ420992 | ||
| vag8a | 6 | |||
| vag8-1 | 17 (1) | U90124 | ||
| vag8-2 | 83 (5) | AJ420993 | ||
| bipAa | 6 | |||
| bipA1 | 83 (5) | P. Cotter | ||
| bipA2 | 17 (1) | This study |
Genes sequenced completely.
prn region 1 was sequenced for all strains, and region 2 was sequenced for 20% of the strains.
The fhaB gene from 13 strains was sequenced, between bases 1 and 6612. This is the region that codes for the secreted, processed, molecule incorporated in ACVs. For the remaining strains, the region comprising bases 2250 to 2750 and containing the polymorphic locus was sequenced.
The tcfA gene from five strains was sequenced completely. Subsequently, the region between bases 395 and 945 was sequenced for all strains. The region between bases 1 and 394 was sequenced for one of five Dutch strains and for all strains from outside The Netherlands.
FIG. 1.
Alleles of genes coding for B. pertussis surface proteins. Numbers refer to the underlined residues. Numbering is relative to the start of the open reading frame. Dots indicate identity, and codons are separated by dashes. Deletions are indicated by an “X.” In bipA “[NNN]360” indicates a region of 1,080 bp that is absent in bipA2. This region contains four repeat regions indicated by arrows. The fifth repeat region, indicated by the arrow, is found in both alleles. In tcfA1, “[NNN]25” indicates a region of 75 bp that is absent from the remaining alleles. A 60-bp segment, flanked by two direct repeats (underlined), is found in tcfA1. The left repeat is found in the alleles tcfA2-5. The arrow below tcfA5 indicates an inserted G, which changes the reading frame and results in premature translational termination two codons downstream. In prn, polymorphism is mainly found in region 1 comprised of 15-bp repeats. Complete sequences can be extracted from the EMBL/GenBank database. Some data were published previously (26).
The genes ptxS1, ptxS3, prn, and tcfA revealed allelic variation in recent isolates (i.e., from the period 1980 to 1999) and were selected for further analyses by using isolates from widely separated geographic regions. Polymorphism in ptxS3 and tcfA was not described previously. For ptxS3, two alleles were found, which differed in a single nucleotide resulting in a silent mutation (Fig. 1). Four tcfA alleles were found (Fig. 1). In total, four polymorphic loci were observed in tcfA. In tcfA1 a 60-bp segment flanked by two 15-bp fragments was present. In the other three alleles, the 60-bp fragment and one repeat was missing, suggesting that a 75-bp deletion had occurred by recombination between the repeats. Two polymorphic sites comprised single base substitutions resulting in amino acid changes. The fourth polymorphic site comprised a homopolymeric tract of nine (tcfa1-3) or ten (tcfA5) Gs. The additional G in tcfA5 results in a frameshift and premature termination of translation.
MLSTs.
Three genes—ptxS1, ptxS3, and tcfA—were used to define MLSTs (Table 2). Although prn also showed polymorphism, it was not used for the MLST typing scheme, since variation in prn is mainly due to insertion and deletion of repeat units, a process that is expected to occur relatively frequently and is reversible. This is in contrast to the (point) mutations observed in ptxS1, ptxS3, and tcfA. In all, nine MLSTs could be distinguished.
TABLE 2.
Global distribution and frequencies of the MLSTs in the period from 1990 to 1999a
| MLST | Alleles | % Distribution by country and periodb
|
No. of isolates (n = 164) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| The Netherlands
|
United States
|
Finland, 90-99 (n = 27) | Italy, 90-99 (n = 10) | Japan, 90-99 (n = 13) | |||||
| Prevac (n = 11) | 90-99 (n = 85) | Prevac (n = 8) | 90-99 (n = 10) | ||||||
| MLST-1 | ptxS1D, ptxS3A, tcfA2 | 45 | 1 | 25 | 8 | ||||
| MLST-2 | ptxS1B, ptxS3A, tcfA2 | 55 | 4 | 50 | 10 | 69 | 23 | ||
| MLST-3 | ptxS1A, ptxS3A, tcfA2 | 25 | 10 | 89 | 40 | 15 | 52 | ||
| MLST-4 | ptxS1A, ptxS3A, tcfA3 | 32 | 11 | 60 | 36 | ||||
| MLST-5 | ptxS1A, ptxS3B, tcfA2 | 38 | 60 | 15 | 41 | ||||
| MLST-7 | ptxS1A, ptxS3A, tcfA5 | 10 | 1 | ||||||
| MLST-8 | ptxS1B, ptxS3B, tcfA2 | 10 | 1 | ||||||
| MLST-9 | ptxS1E, ptxS3A, tcfA1 | 25 | 2 | ||||||
For the Netherlands and the United States strains from the prevaccination period have been included.
Prevac, prevaccination period; 90-99, period from 1990 to 1999. n, number of isolates.
Temporal trends in MLST frequencies in The Netherlands.
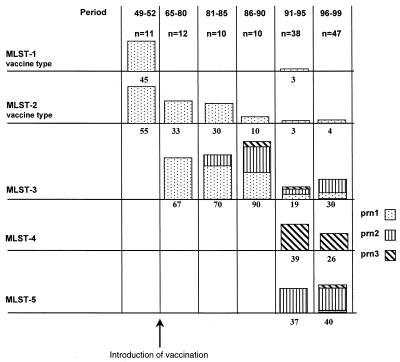
The availability of an extensive, well-defined strain collection allowed a detailed analysis of MLST frequencies in The Netherlands. To investigate temporal trends, strains were stratified in several different periods: 1949 to 1952 (the prevaccination period), 1965 to 1980, 1981 to 1985, 1986 to 1990, 1991 to 1995, and 1996 to 1999 (Fig. 2). Two MLSTs (MLST-1 and MLST-2) were observed in the prevaccination era, comprising 45 and 55% of the population, respectively. The two Dutch vaccine strains each represent one of the two MLSTs. MLST-1 disappeared after the introduction of vaccination and was only observed again in the period from 1991 to 1995, when it comprised 3% of the strains analyzed. The frequency of MLST-2 also decreased after the introduction of vaccination, dropping to 33, 30, 10, 3, and 4%, respectively, in the periods 1965 to 1980, 1981 to 1985, 1986 to 1990, 1991 to 1995, and 1996 to 1999. In the period from 1965 to 1980, a novel MLST was found (MLST-3), initially comprising 67% of the isolates, and increasing in frequency to 70 and 90% in the periods from 1980 to 1985 and 1986 to 1990, respectively. In the subsequent two periods, 1991 to 1995 and 1996 to 1999, the frequency of MLST-3 dropped to 19 and 30%, respectively. In the period from 1991 to 1995, two new MLSTs (MLST-4 and MLST-5) emerged, which were found at frequencies of 39 and 37%, respectively, in the period from 1991 to 1995 and of 26 and 40% in the period from 1996 to 1999.
FIG. 2.
MLST frequencies in The Netherlands in the period from 1949 to 1999. Frequencies of MLSTs within the investigated periods are indicated by bars, and percentages are given below the bars. The distribution of prn alleles within a particular MLST is indicated by distinct segments.
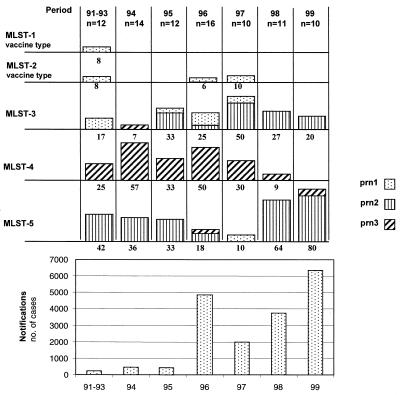
In 1996 a severe pertussis epidemic occurred in The Netherlands, and the incidence has remained high ever since (7) (Fig. 3). To determine whether the upsurge could be linked to particular MLSTs, the period from 1991 to 1999 was studied in more detail. Isolates were stratified in 12-month periods from May to April of the following year. These periods encompass the annual epidemic peak that spans the calendar year. Only a few isolates from the years 1991, 1992, and 1993 were available, and therefore they were pooled (Fig. 3). MLST-4 was observed to dominate in the years 1994 and 1996, comprising 57 and 50% of the isolates, respectively. In the years from 1991 to 1993, 1998, and 1999, the majority of isolates were represented by MLST-5. In the last 3 years analyzed, MLST-5 expanded in frequency from 10 to 64 and 80%.
FIG. 3.
MLST frequencies and pertussis notifications in The Netherlands in the period from 1991 to 1999. Pertussis notifications are from the period May to April. See the legend to Fig. 2 for further details.
Global distribution of MLSTs.
For a comparison of the global distribution of MLSTs, isolates from Finland, Italy, Japan, The Netherlands, and the United States were compared (Table 2). Isolates were stratified in two periods: the prevaccination period and the period from 1990 to 1999. Strains from the prevaccination period were only available from The Netherlands and the United States. As mentioned previously, MLST-1 and MLST-2 predominated in The Netherlands in the prevaccination period (frequencies of 45 and 55%, respectively). These MLSTs were also observed in high frequencies in the prevaccination era in the United States, when they comprised 25 and 50% of the isolates. In the United States, an additional MLST was found (MLST-9) in this period that was associated with 25% of the isolates. Although predominant in the prevaccination era, all three MLSTs were found at a low frequency (0 to 10%) in the period from 1990 to 1999 in all countries, except for Japan where MLST-2 comprised 69% of the isolates. In the period from 1990 to 1999, seven MLSTs were found in the five countries compared. The frequencies of the MLSTs differed between countries. MLST-4 dominated in Italy (frequency of 60%), MLST-3 dominated in Finland (frequency of 89%), while MLST-5 dominated in the United States (frequency of 60%). MLST-4 and MLST-5 were found in approximately equal frequencies (32 and 38%) in The Netherlands in this period.
Linkage between MLSTs and pertactin alleles.
For MLSTs that were observed ≥3 times in a country, linkage with prn alleles was determined in the period from 1990 to 1999 (Table 3). A nonrandom association between MLSTs and prn alleles was observed (P < 0.0001). MLST-4 was associated with a single prn allele (prn3). MLST-2 was linked with prn1 (92%) and prn2 (8%). MLST-3 was linked with prn1-5, whereas linkage with prn2 was most frequent (67%). Finally, MLST-5 was associated with prn1-3, with linkage with prn2 (92%) being most frequent.
TABLE 3.
Linkage between MLSTs and prn alleles in the period from 1990 to 1999a
| MLST | Country | % prn alleles (no. of isolates analyzed)
|
No. of isolates | ||||
|---|---|---|---|---|---|---|---|
| prn1 | prn2 | prn3 | prn4 | prn5 | |||
| MLST-2 | The Netherlands | 100 (3) | 3 | ||||
| Japan | 89 (8) | 11 (1) | 9 | ||||
| Total | 92 (11) | 8 (1) | 12 | ||||
| MLST-3 | The Netherlands | 35 (7) | 61 (13) | 4 (1) | 21 | ||
| Finland | 4 (1) | 84 (20) | 8 (2) | 4 (1) | 24 | ||
| Italy | 25 (1) | 75 (3) | 4 | ||||
| Total | 16 (8) | 67 (33) | 8 (4) | 2 (1) | 6 (3) | 49 | |
| MLST-4 | The Netherlands | 100 (27) | 27 | ||||
| Finland | 100 (3) | 3 | |||||
| Italy | 100 (6) | 6 | |||||
| Total | 100 (36) | 36 | |||||
| MLST-5 | The Netherlands | 3 (1) | 93 (30) | 5 (2) | 33 | ||
| United States | 100 (6) | 6 | |||||
| Total | 3 (1) | 92 (36) | 5 (2) | 39 | |||
Linkage was determined for MLSTs which were identified ≥3 times in a country. The association between MLST and prn alleles was nonrandom (χ2 test, P < 0.0001). Numbers of isolates analyzed are indicated in parentheses.
DISCUSSION
Initially, we attempted to find polymorphism in B. pertussis housekeeping genes. The housekeeping genes glyA, Idh, lap, pgm, and got from 4 to 11 strains were analyzed. Polymorphism was only observed in adk and got, which both occurred as two alleles differing in one base (unpublished data). In view of the restricted polymorphism found in B. pertussis housekeeping genes (34; data not shown), we attempted to increase the likelihood of identifying allelic variation by analysis of genes coding for surface proteins, which may be subject to more intensive selective pressures. Our data provide the first extensive analysis of polymorphism in all five components used for pertussis ACVs. We confirmed previous observations that PtxS1 and Prn were polymorphic (4, 24-26). Interestingly, although PtxS1 was polymorphic, the other four protein subunits of pertussis toxin were monomorphic. Only one, silent, mutation was detected in the gene for PtxS3. This suggests that the S1 subunit is more immunogenic compared to the other Ptx subunits, and/or that antibodies to PtxS1 affect strain fitness more severely. As to the remaining components of ACVs, a single nonsilent mutation was observed in the genes for FHA and Fim2, while the gene for Fim3 was monomorphic. All Dutch isolates from 1965 or later harbored the fhaB1 and fim2-1 alleles. In conclusion, of the five proteins included in pertussis ACVs, evidence for a mismatch between vaccine strains and clinical isolates was only found for Ptx and Prn (4, 15, 24-26). An important question is whether the observed mismatches affect the efficacy of pertussis vaccines. Using a mouse model, we have found that variation in Prn affects the efficacy of a WCV (21). One may argue that, since ACVs induce higher titers against Ptx and Prn compared to WCV, they may be less affected by the mismatch. On the other hand, protection conferred by ACVs is based on only a few proteins, whereas WCV induces a much broader immunity based on antigens (such as lipopolysaccharide), which are highly conserved. Both types of vaccines are now used, and a comparison of countries by using either a WCV or an ACV may resolve this issue in the future.
We also studied polymorphism in a number of surface-associated proteins, which are not found in ACVs: BipA, BrkA, TcfA, OmpP, OmpQ, and Vag8. No polymorphism was detected in brkA and ompP and, with the exception of tcfA, polymorphism in the remaining genes was limited. Only two alleles were observed for bipA, ompQ, and vag8, only one of which was found in the period from 1965 to 1999 in The Netherlands. The lack of polymorphism in the selected surface proteins was unexpected, since some of them have been implicated in immunity. For example, micelles containing TcfA and Vag8 were protective in the intracerebral mouse test if low, nonprotective amounts of Ptx were added (18). OmpP is the major outer membrane protein of B. pertussis (2). High titers of bactericidal antibodies directed against OmpP were induced after vaccination with micelles in mice, suggesting it may represent an effective vaccine component (28). The monomorphic nature of B. pertussis OmpP is unlikely to be due to structural constraints, since 0.4 to 1% divergence was observed with its B. bronchiseptica and B. parapertussis homologues (data not shown).
Four tcfA alleles were identified. The tcfA1 allele contains a 60-bp segment flanked by two direct repeats. The 60-bp segment and one repeat was not found in tcfA2, tcfA3, and tcfA5, and it seems likely that it was deleted by recombination between the repeats. Since deletion of a sequence between two direct repeats is a more likely event than a duplication of a nonrepeated sequence, tcfA1 is probably the progenitor of tcfA2, tcfA3, and tcfA5. Interestingly, tcfA1 is found in the 18323 strain, an atypical B. pertussis strain sometimes erroneously classified more closely to B. bronchiseptica than to B. pertussis. We speculate that the 18323 strain may be closely related to the original B. bronchiseptica strain, from which B. pertussis evolved (27, 34). Homologues of tcfA were also found in the B. bronchiseptica and B. parapertussis genomes (www.sanger.ac.uk). This finding is consistent with the observation of Finn et al. that a B. pertussis tcfA probe hybridized to DNA from B. parapertussis and B. bronchiseptica (14). The tcfA5 allele contained a frameshift mutation in a homopolymeric G-track, suggesting that it is subject to phase variation. Homopolymeric G- or C-tracks have been shown to be involved in fimbrial phase variation at the transcriptional level in B. pertussis (38).
We sequenced 27,862 bases derived from 15 gene segments. In all, we found 22 point mutations and four insertions or deletions in 27 polymorphic loci, indicating a very low degree of polymorphism in B. pertussis. For Mycobacterium tuberculosis, also a very homogeneous species, the lack of diversity has been suggested to be due to an evolutionary recent origin and worldwide spread of a clone in an episode of periodic selection (31). It is presumed that the agent of tuberculosis arose from the very closely related cattle pathogen Mycobacterium bovis by host specialization occurring since the domestication of this animal some 8,000 to 10,000 years ago (20). In a similar vain, the lack of allelic polymorphism in B. pertussis is consistent with this species evolving from a subset of B. bronchiseptica, which has adapted to the human host relatively recently (27, 34).
The virtual absence of silent mutations in B. pertussis suggests that most amino acid variations observed increase strain fitness. The lack of polymorphism in B. pertussis surface-associated proteins in recent clinical isolates underlines the significance of the variation found in Prn, PtxS1, and TcfA. The polymorphic loci in these proteins may interact directly with the immune system or other host targets, such as receptors used for attachment. In the period from 1990 to 1999, most variation was observed in Prn and TcfA. Many studies have shown the important role of Prn in protective immunity (5, 6, 33). We have found that, in a mouse model, the Dutch whole-cell vaccine is less effective against strains that produce nonvaccine Prn types (21). Our study suggests that also antibodies to TcfA may be effective in conferring protection.
MLST has proven to be useful for studying the epidemiology of bacterial pathogens (10, 12, 19), and we categorized B. pertussis isolates in MLSTs, based on polymorphism in ptxS1, ptxS3, and tcfA. Prn was not included in the MLST scheme, since variation is based on recombination within the repeated region or slipped-strand mispairing, phenomena, which occur at relatively high frequency. Interestingly, the prn alleles were not distributed randomly over the MLSTs (P < 0.0001). A nonrandom distribution of prn alleles among PFGE types was observed independently by two groups (4, 37). Focusing on the three MLSTs, which comprise a sufficient number of analyzed strains (i.e., 36 to 49), the following can be observed. MLST-3 and MLST-5 were associated with five and three prn alleles, respectively (Table 3). However, prn2 predominated within these MLSTs (frequencies of 67 and 92%, respectively). In contrast, MLST-4 was found to be associated with the prn3 allele only. Gupta et al. have shown that pathogen populations may segregate into discrete strains with nonoverlapping antigenic structures if these structures are immunodominant (16). Thus, the nonrandom association of prn alleles with MLSTs may reflect strong immune selection focused on Prn and other (as-yet-unknown) B. pertussis antigens.
In accordance with our previous studies based on IS1002 restriction fragment length polymorphism, MLST analysis revealed large changes in the bacterial population subsequent to the introduction of the WCV in The Netherlands and the United States (4, 35, 36). MLST-1 and MLST-2, types to which the two Dutch vaccine strains belong, were predominant in the prevaccination period but only detected at a very low frequency in the B. pertussis population in subsequent periods. A plausible explanation for this phenomenon is that vaccination has shifted the competitive balance between strains and selected for variants less affected by vaccine-induced immunity. In 1996 there was a severe epidemic of pertussis in The Netherlands, and the incidence has remained high ever since (7, 8). MLST did not reveal obvious differences between the bacterial populations from 1991 to 1995 and from 1996 to 1999, respectively. This may be due to lack of appropriate markers in our MLST system. Alternatively, the relationship between epidemics and bacterial strains may be complex and involve factors unrelated to the bacterial population, such as changes in surveillance, host immunity, and vaccine quality. However, a more detailed analysis of the period from 1991 to 1999 did reveal interesting trends. The twofold increase in notifications in 1994 compared to 1991 to 1993 was associated with an increase in the frequency of MLST-4 of from 25 to 57%. The 10-fold increase in notifications in 1996 compared to 1995 was associated with an increase in frequency of MSLT-4 from 33 to 50%. Also in 1998 and 1999 there was a substantial increase in notifications compared to 1997 (1.9- and 3.2-fold, respectively) associated with an expansion of MLST-5 from 10% in 1997 to 64 and 80% in 1998 and 1999, respectively. This suggests that epidemic years are associated with clonal expansion of particular strains. The expansion of MLST-5 is particularly remarkable. MLST-5 is identical to MLST-3 except for a silent mutation in ptxS3, and whereas MLST-5 increased in frequency from 10% in 1997 to 80% in 1999 the frequency of MLST-3 was reduced from 50 to 20% in the same period. Since the mutation in ptxS3 is very likely neutral, the distinctive behavior of these two MLSTs may be due to an as-yet-unidentified polymorphic gene, which has a strong effect on strain fitness. We are currently trying to identify this gene. The identification of strains, which expand in epidemic years, illustrates the value of MLST analyses.
We extended our MLST analysis to include a number of widely separated geographic regions: Finland, Italy, Japan, and the United States. MLST-4 and MLST-5 were found to dominate in Italy and the United States, respectively (Table 2). Significantly, these MLSTs predominated in years with the highest pertussis notifications in The Netherlands. However, in contrast to The Netherlands, where both MLSTs were present in approximately equal frequencies in the period from 1990 to 1999 (32 and 38%, respectively), only one type predominated in Italy and the United States. In Finland a third MLST (MLST-3) was found to predominate, comprising 89% of the isolates. In Japan, MLST-2 comprised 69% of the isolates. Thus, although each region showed distinctive MLST frequencies, in three of the five regions MLST-4 and MLST-5 were predominant. Of the three regions in which MLST-4 and MLST-5 predominate, two (The Netherlands and the United States) have reported an increased pertussis incidence (3, 7, 8, 17). MLST-4 and MLST-5 may represent newly emerged, successful clones. Further analysis of strains belonging to MLST-4 and MLST-5 may reveal why B. pertussis has remained such a successful pathogen despite intensive vaccination.
Acknowledgments
This study was supported by grant 247221 from the Ministry of Public Health, Welfare, and Sport (VWS).
REFERENCES
- 1.Andrews, R., A. Herceg, and C. Roberts. 1997. Pertussis notifications in Australia, 1991 to 1997. Commun. Dis. Intell. 21:145-148. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. K., T. R. Parr, Jr., C. D. Parker, and R. E. Hancock. 1986. Bordetella pertussis major outer membrane porin protein forms small, anion-selective channels in lipid bilayer membranes. J. Bacteriol. 166:212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, J. W., and R. R. Wittler. 1994. Return of epidemic pertussis in the United States. Pediatr. Infect. Dis. J. 13:343-345. [DOI] [PubMed] [Google Scholar]
- 4.Cassiday, P., G. Sanden, K. Heuvelman, F. Mooi, K. M. Bisgard, and T. Popovic. 2000. Polymorphism in Bordetella pertussis pertactin and pertussis toxin virulence factors in the United States, 1935-1999. J. Infect. Dis. 182:1402-1408. [DOI] [PubMed] [Google Scholar]
- 5.Charles, I. G., J. L. Li, M. Roberts, K. Beesley, M. Romanos, D. J. Pickard, M. Francis, D. Campbell, G. Dougan, M. J. Brennan, C. R. Manclark, M. A. Jensen, I. Heron, A. Chubb, P. Novotny, and N. F. Fairweather. 1991. Identification and characterization of a protective immunodominant B-cell epitope of pertactin (P.69) from Bordetella pertussis. Eur. J. Immunol. 21:1147-1153. [DOI] [PubMed] [Google Scholar]
- 6.Cherry, J. D., J. Gornbein, U. Heininger, K. Stehr, J. Storsaeter, H. O. Hallander, L. Gustafsson, P. Olin, I. JabbalGill, A. N. Fisher, R. Rappuoli, S. S. Davis, and L. Illum. 1998. A search for serologic correlates of immunity to Bordetella pertussis cough illnesses. Vaccine 16:2039-2046. [DOI] [PubMed] [Google Scholar]
- 7.de Melker, H. E., M. A. Conyn van Spaendonck, H. C. Rumke, J. K. van Wijngaarden, F. R. Mooi, and J. F. Schellekens. 1997. Pertussis in The Netherlands: an outbreak despite high levels of immunization with whole-cell vaccine. Emerg. Infect. Dis. 3:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Melker, H. E., J. F. Schellekens, S. E. Neppelenbroek, F. R. Mooi, H. C. Rumke, and M. A. Conyn-van Spaendonck. 2000. Reemergence of pertussis in the highly vaccinated population of The Netherlands: observations on surveillance data. Emerg. Infect. Dis. 6:348-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Serres, G., N. Boulianne, M. Douville Fradet, and B. Duval. 1995. Pertussis in Quebec: ongoing epidemic since the late 1980s. Can. Commun. Dis. Rep. 21:45-48. [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 12.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn, T. M., and D. F. Amsbaugh. 1998. Vag8, a Bordetella pertussis Bvg-regulated protein. Infect. Immun. 66:3985-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn, T. M., and L. A. Stevens. 1995. Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol. Microbiol. 16:625-634. [DOI] [PubMed] [Google Scholar]
- 15.Guiso, N., C. Boursaux-Eude, C. Weber, S. Z. Hausman, H. Sato, M. Iwaki, K. Kamachi, T. Konda, and D. L. Burns. 2001. Analysis of Bordetella pertussis isolates collected in Japan before and after introduction of acellular pertussis vaccines. Vaccine 19:3248-3252. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, S., M. C. J. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2:437-442. [DOI] [PubMed] [Google Scholar]
- 17.Guris, D., P. M. Strebel, B. Bardenheier, M. Brennan, R. Tachdjian, E. Finch, M. Wharton, and J. R. Livengood. 1999. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin. Infect. Dis. 28:1230-1237. [DOI] [PubMed] [Google Scholar]
- 18.Hamstra, H. J., B. Kuipers, D. Schijfevers, H. G. Loggen, and J. T. Poolman. 1995. The purification and protective capacity of Bordetella pertussis outer membrane proteins. Vaccine 13:747-752. [DOI] [PubMed] [Google Scholar]
- 19.Hoe, N., K. Nakashima, D. Grigsby, X. Pan, S. J. Dou, S. Naidich, M. Garcia, E. Kahn, D. Bergmire-Sweat, and J. M. Musser. 1999. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg. Infect. Dis. 5:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur, V., T. S. Whittam, and J. M. Musser. 1994. Is Mycobacterium tuberculosis 15,000 years old? J. Infect. Dis. 170:1348-1349. [DOI] [PubMed] [Google Scholar]
- 21.King, A. J., B. Berbers, H. F. L. M. Van Oirschot, P. Hoogerhout, K. Knipping, and F. R. Mooi. 2001. Role of the polymorphic region 1 of the Bordetella pertussis protein pertactin in immunity. Microbiology 147:2885-2895. [DOI] [PubMed] [Google Scholar]
- 22.Li, Z. M., J. H. Hannah, S. Stibitz, N. Y. Nguyen, C. R. Manclark, and M. J. Brennan. 1991. Cloning and sequencing of the structural gene for the porin protein of Bordetella pertussis. Mol. Microbiol. 5:1649-1656. [DOI] [PubMed] [Google Scholar]
- 23.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mastrantonio, P., P. Spigaglia, H. van Oirschot, H. G. van der Heide, K. Heuvelman, P. Stefanelli, and F. R. Mooi. 1999. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology 145:2069-2075. [DOI] [PubMed] [Google Scholar]
- 25.Mooi, F. R., Q. He, H. van Oirschot, and J. Mertsola. 1999. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect. Immun. 67:3133-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. J. van der Heide, W. Gaastra, and R. J. L. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musser, J. M., E. L. Hewlett, M. S. Peppler, and R. K. Selander. 1986. Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poolman, J., H. J. Hamstra, A. Barlow, B. Kuipers, H. Loggen, and J. Nagel. 1990. Outer membrane vesicles of Bordetella pertussis are protective antigens in the mouse intracerebral challenge model, p. 202-206. In Proceedings of the Sixth International Symposium on Pertussis. DHHS publication no. (FDA) 90-1164. Department of Health and Human Services, Bethesda, Md.
- 29.Rambow, A. A., R. C. Fernandez, and A. A. Weiss. 1998. Characterization of Brka expression in Bordetella bronchiseptica. Infect. Immun. 66:3978-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spratt, B. G., and M. C. Maiden. 1999. Bacterial population genetics, evolution and epidemiology. Philos. Trans. R. Soc. London B. Biol. Sci. 354:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockbauer, K. E., B. Fuchslocher, J. F. Miller, and P. A. Cotter. 2001. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 39:65-78. [DOI] [PubMed] [Google Scholar]
- 33.Storsaeter, J., H. O. Hallander, L. Gustafsson, P. Olin, I. JabbalGill, A. N. Fisher, R. Rappuoli, S. S. Davis, and L. Illum. 1998. Levels of anti-pertussis antibodies related to protection after household exposure to Bordetella pertussis. Vaccine 16:1907-1916. [DOI] [PubMed] [Google Scholar]
- 34.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Zee, A., S. Vernooij, M. Peeters, J. van Embden, and F. R. Mooi. 1996. Dynamics of the population structure of Bordetella pertussis as measured by IS1002-associated RFLP: comparison of pre- and post-vaccination strains and global distribution. Microbiology 142:3479-3485. [DOI] [PubMed] [Google Scholar]
- 36.van Loo, I. H., H. G. van der Heide, N. J. Nagelkerke, J. Verhoef, and F. R. Mooi. 1999. Temporal trends in the population structure of Bordetella pertussis during 1949-1996 in a highly vaccinated population. J. Infect. Dis. 179:915-923. [DOI] [PubMed] [Google Scholar]
- 37.Weber, C., C. Boursaux-Eude, G. Coralie, V. Caro, and N. Guiso. 2001. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J. Clin. Microbiol. 39:4396-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems, R., A. Paul, H. G. van der Heide, A. R. ter Avest, and F. R. Mooi. 1990. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO. J. 9:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]