Abstract
We investigated the influence of telomere proximity and composition on the expression of an EGFP reporter gene in human cells. In transient transfection assays, telomeric DNA does not repress EGFP but rather slightly increases its expression. In contrast, in stable cell lines, the same reporter construct is repressed when inserted at a subtelomeric location. The telomeric repression is transiently alleviated by increasing the dosage of the TTAGGG repeat factor 1 (TRF1). Upon a prolongated treatment with trichostatin A, the derepression of the subtelomeric reporter gene correlates with the delocalization of HP1α and HP1β. In contrast, treating the cells with 5 azacytidin, a demethylating agent, or with sirtinol, an inhibitor of the Sir2 family of deacetylase, has no apparent effect on telomeric repression. Overall, position effects at human chromosome ends are dependent on a specific higher-order organization of the telomeric chromatin. The possible involvement of HP1 isoforms is discussed.
Introduction
Telomeres have a structure that allows the cell's DNA repair machinery to distinguish natural chromosome ends from 'broken' DNA ends (Lundblad, 2000). They also provide a means for the complete replication of the chromosomal DNA (Blackburn, 2000). Furthermore, the structure and spatial localization of telomeric chromatin play an important role in the nuclear compartmentalization of gene expression and probably of other chromosomal transactions, such as replication initiation, condensation, segregation, recombination and repair (Gilson et al., 1993). These properties are usually revealed by telomeric position effect (TPE). For instance, telomere proximity induces gene silencing and late activation of replication origins (Gottschling et al., 1990; Ferguson and Fangman, 1992; Nimmo et al., 1994; Rudenko et al., 1995; Cryderman et al., 1999).
The heterochromatic nature of mammalian telomeres and their capacity to induce position effect have been controversial for many years (Bayne et al., 1994; Sprung et al., 1996; Ofir et al., 1999; Smith and Higgs, 1999; Wright et al., 1999; Baur et al., 2001). Until now, the best evidence for human TPE (hTPE) consists in the delayed replication timing induced by a chromosome truncation (Ofir et al., 1999) and in the telomere length dependency of the expression of a subtelomeric reporter gene (Baur et al., 2001). In diseases involving telomeric rearrangements, a modulation of hTPE might lead to inappropriate gene expression, as in the myopathy FSHD (Gabellini et al., 2002).
We investigated the influence of telomere proximity and composition on the expression of an EGFP reporter gene in human cells. Our findings demonstrate that TPE in human cells is dependent on a specific higher-order organization of the telomeric chromatin.
Results and discussion
The proximity of telomeric DNA activates gene expression in transient assays
We first asked whether a stretch of telomeric DNA could act as a cis-acting regulatory element in transient transfection assays. For this purpose, C33-A cells (a cervical undifferentiated carcinoma line) were transfected with increasing amounts of plasmid DNAs bearing an EGFP gene under the control of the CMV promoter with or without 1.6 kb of adjacent TTAGGG repeats (pCMVTelo and pCMV, respectively; Figure 1A). The molar concentration of the transfected CMV promoter DNA was maintained constant by adding an appropriate amount of plasmid containing only the CMV promoter DNA. Putative variations in transfection efficiency were evaluated by co-transfecting with pBLCat DNA (Waltzer et al., 1996) and by normalizing the percentage of EGFP-expressing cells to the amount of CAT expression.
Figure 1.
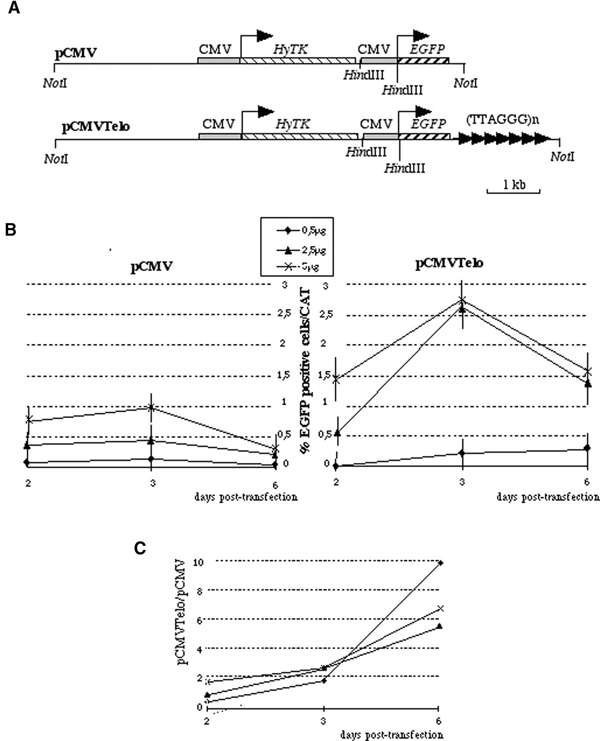
Transient expression assays reveals a transactivation activity of telomeric repeats. (A) Schematic map of the two reporter plasmids used in this study. pCMVTelo contains a stretch of 1.6 kb TTAGGG in close proximity to the reporter EGFP gene driven by a CMV promoter. At 1.8 kb from the TTAGGG repeats, we introduced the fusion gene between hygromycin phosphotransferase and HSV1 thymidine kinase (HyTK) conferring the resistance to hygromycin as a selectable marker. The control vector lacking the telomeric 1.6 kb stretch was named pCMV. (B) Transient expression assays with various amount of pCMV or pCMVTelo circular DNA. A time course of the EGFP expression after transfection was determined. The percentage of EGFP-positive cells is corrected for transfection efficiency determined by CAT assay. The values correspond to the average of at least three independent experiments. We estimated the standard error to be <20%. (C) Ratio of the percentage of EGFP-positive cells in pCMVTelo transfection to that in pCMV transfections.
An enhanced expression of EGFP correlates with the dosage of the plasmid DNA and peaks 3 days after transfection (Figure 1B). The increase in the percentage of EGFP-positive cells is much more pronounced with pCMVTelo than with pCMV (Figure 1B and C). Therefore, TTAGGG repeats do not exhibit silencing properties in transient transfection assays. Thus, it appears unlikely that hTPE results simply from the binding of a transcriptional repressor to telomeric DNA.
Repressive effects of telomere proximity in stably transfected cells
In order to test whether the chromosomal context is important to uncover the repressive properties of telomeric DNA, we integrated the same EGFP reporter cassette at the immediate proximity of a telomere. Since cloned human telomeric DNA can seed the formation of new telomeres (Farr et al., 1991; Hanish et al., 1994), linearized pCMVTelo DNA was transfected into C33-A cells. Transfections with linearized pCMV DNA was used as a negative control. Figure 2 shows that, in three independent transfections (1, 2 and 3), within the first few days, the percentage of EGFP-positive cells is significantly higher among pCMVTelo than among pCMV transfectants. This effect is more pronounced for transfection 1, but it can clearly be seen in 2 and 3 (see the enlarged views in Figure 2). This transactivation of the telomeric DNA-linked reporter at an early stage of the transfection process is in agreement with the results obtained in transient transfection with circular DNA (Figure 1).
Figure 2.

Telomeric silencing in populations of transfected cells. The pCMV and pCMVTelo DNA were linearized with NotI (Figure 1A) and used to transform C33-A cells. By co-transfection with pBLCat, we estimated that the efficiencies were roughly similar in all situations (data not shown). The percentage of EGFP-positive cells was analyzed by Facscan. Three independent transfections are shown (1, 2 and 3). Three days after transfection, hygromycin (400 μg/ml) was added. The boxes show an enlarged view of the beginning of the kinetics for 2 and 3.
From the three transfections shown in Figure 2, several clones were established from hygromycin resistant cells. Successful telomere seeding in pCMVTelo clones was confirmed by detection of a diffuse hybridization signal using the EGFP probe (Hanish et al., 1994). Sixteen out of 18 analyzed clones correspond to a telomeric fragmentation (data not shown). For three out of three randomly chosen telomeric clones, the presence of the EGFP DNA at one chromosome end was confirmed on metaphase spreads by fluorescence in situ hybridization (FISH), using a pCMV DNA probe (Figure 3A; data not shown). These data reveal a very high seeding efficiency for C33-A cells, indicating that the population of pCMVTelo-transfected cells is likely to contain a large majority of telomeric integration sites, probably at different chromosome ends.
Figure 3.

Telomeric silencing in clones. (A) Localization of the EGFP gene at 16p by chromosome 16 painting (image a) and FISH with an EGFP probe (image b); the position of 16p is marked by arrows. (B) The percentage of EGFP-positive cells in clones presenting a single insertion of the reporter gene. These clones were obtained from three independent transfections with either pCMV or PCMVTelo. (C) The percentage of EGFP-positive cells plotted versus the length of the EGFP-linked telomere [eTRF in kb of (TTAGGG)n]. The eTRF value was determined by Southern blotting after probing HindIII-cut genomic DNA with an EGFP probe.
After 10 days in hygromycin, the percentage of EGFP-positive cells among the population of transfected cells becomes higher with pCMV than with pCMVTelo, reversing the transactivation effect observed with pCMVTelo-transfected cells at the beginning of the kinetics (Figure 2). This striking change is likely to coincide with the integration of the plasmid DNA into chromosome and the seeding event in pCMVTelo-transfected cells. With time, the percentage of positive pCMV transfectants increases progressively, whereas the rate of positive cells after transfection with pCMVTelo plateaus at ∼10%.
Starting from the cells transfected with pCMVTelo, sorted populations either enriched or depleted in EGFP-positive cells were cultured for ∼20 mean population doublings (MPDs). The EGFP expression remained relatively stable, whether or not the medium contains hygromycin (data not shown). Thus, the percentage of EGFP-positive cells is a long-term characteristic of a population of cells. Furthermore, this indicates that the selective pressure exerted by hygromycin on the expression of the HyTK gene does not greatly influence the expression of EGFP, at least for 20 MPDs. This uncoupling between the HyTK and the EGFP expression might be attributed to a putative boundary activity of the CMV promoter.
Only clones containing a single copy of the EGFP gene were studied further. Single-copy insertions were revealed as an equal signal intensity between a pCMV plasmid and a β-actin probe by Southern blotting (data not shown). By these criteria, all 16 telomeric clones and 10 out of 20 analyzed pCMV clones exhibit a single integrated copy of the plasmid DNA. Both the pCMV and the pCMVTelo clones are mosaics of EGFP-positive and -negative cells (Figure 3B). For all these clones, a population deprived of EGFP-positive cells was sorted and cultured for 3 months without any significant change in EGFP expression (data not shown). This indicates that the repression of EGFP is not spontaneously reversible within the time limits of this experiment. For pCMV clones, the rate of positive cells within a positive clone ranges from 0.02 to 70%. In contrast, the pCMVTelo clones have an expression efficiency ranging from 0.1 to 3% in positive clones (Figure 3B). The fact that the range of EGFP-expressing cells is more constrained when the transgene is juxtaposed to a telomere than when it is internally inserted indicates that telomere proximity confers a characteristic position effect.
TPE correlates with telomeric protein composition
We plotted the percentage of EGFP-positive cells corresponding to the different pCMVTelo clones as a function of the median length of the associated telomeric DNA (EGFP telomeric restriction fragment, eTRF; Figure 3C). There is no obvious correlation between the two values (Figure 3B), in agreement with previous studies (Sprung et al., 1996). This seemingly contrasts with the fact that a telomere overelongation of 9 kb correlates with an increased TPE (Baur et al., 2001). This discrepancy can be explained by cell type differences and/or the range of telomere length variation. Alternatively, what controls hTPE might not be the telomere length per se but accompanied changes in chromatin structure.
Therefore, we modified the dosage of a major component of the telomeric chromatin, the TTAGGG repeat factor 1 (TRF1) protein (Chong et al., 1995; Bilaud et al., 1996). We constructed a TRF1 expression vector (pITRF1) where a FLAG-tagged TERF1 gene is co-transcribed with the neomycin phosphotransferase gene. The two open reading frames are co-translated due to the presence of an internal ribosome entry site sequence (IRES). The pattern of TRF1-FLAG expression was roughly similar after transfection of two pCMVTelo clones, named 4C4 and 4C11, or of two pCMV clones, named 2A3 or 3B10. By immunoblotting using anti-FLAG antibodies, TRF1-FLAG reaches a plateau after ∼11 MPDs after selection and then remains relatively constant for at least 17 MPDs (Figure 4A and B for 4C11 and 2A3, respectively; data not shown for 4C4 and 3B10). Importantly, by immunoblotting with anti-TRF1 antibodies, it appears that the total amount of TRF1 protein, i.e. the endogenous one and TRF1-FLAG, is significantly enhanced, compared to pIRES-transfected cells, after 11 MPD days and then remains constant (Figures 4A and B; data not shown). This is in agreement with the kinetics of TRF1-FLAG expression and indicates that the level reached by TRF1-FLAG at the plateau is higher than that of the endogenous TRF1. Although it was previously reported that an increased TRF1 or FLAG-TRF1 dosage decreases telomere length (van Steensel and de Lange, 1997), no apparent change of telomere length was observed for 20 MPDs, both for the EGFP-linked telomere and for the bulk of telomeres (data not shown). More cell divisions might be required to detect a change in telomere length dynamics. Indeed, overexpression of full-length TRF1 leads to a relatively slow rate of shortening, ranging between 3 and 10 nt/MPD (van Steensel and de Lange, 1997), Alternatively, a small subset of cells could express TRF1-FLAG at a very high level, leading to a rapid telomere shortening in only a minority of cells. However, this possibility appears highly unlikely, since immunofluorescence studies with FLAG antibodies revealed that >90% of them expressed TRF1-FLAG at a high level during the time course of the experiments (see Supplementary figure 1 available at EMBO reports Online). Importantly, we have been unable to detect a small fraction of cells expressing a significantly higher amount of TRF1
Figure 4.

Overexpression of TRF1 antagonizes telomeric silencing. The pITRF1 or pIRES plasmid DNAs were transfected in the 3B10 and 2A3 pCMV clones and the 4C4 and 4C11 pCMVTelo clones. Four days after transfection, a neomycin selection was applied (t0) and cells were cultured for >30 MPDs. (A) TRF1 expression was determined in 4C11 cells transfected with pIRES or pITRF1 by immunoblotting using either anti-FLAG antibodies (a monoclonal anti-M2 antibody; Sigma) or anti-TRF1 antibodies. Anti-tubulin antibodies were used to evaluate the loading variations. (B) The same type of immunoblotting experiments as in (A) but with 2A3 transfected cells. (C) The entire population of neomycin resistant clones was analyzed by Facscan for EGFP expression.
Nevertheless, the expression of TRF1-FLAG at 7 MPDs after selection is sufficient to lead to a large increase in the percentage of EGFP-positive 4C4 and 4C11 cells when normalized to the expression of EGFP in cells transfected with the parental DNA (pIRES) (Figure 4C). The level of EGFP-positive cells remains elevated until 20 MPDs, when it returns to base line (Figure 4C). The increase is more pronounced with the 4C11 clone (culminating at ∼8-fold) than with the 4C4 clone (culminating at 3-fold) (Figure 4C). No variation in EGFP expression was monitored when TRF1-FLAG was expressed in 3B10 or 2A3 cells (Figure 4C), indicating that the TRF1 effect is specific for the EGFP gene adjoining a telomere. We conclude that TRF1-FLAG expression correlates with the derepression of a subtelomeric EGFP gene. This establishes that TRF1 is an hTPE-modifier in two different cell lines. However, this does not imply that the effect of TRF1 on hTPE is direct. For instance, a modified dosage of TRF1 might perturb the integrity of telomeric chromatin, altering the binding or titrating out some key TPE players. Finally, it seems unlikely that the re-establishment of TPE corresponds to a counterselection of TRF1-expressing cells because the TRF1 dosage remains unaltered (Figure 4A). Therefore, we favor the idea that the transient derepression reveals an adaptation process of the telomeric chromatin to the new concentration of TRF1, allowing it to regain at least part of its properties.
These results show that hTPE occurs at a seeded telomere in human C33-A cells. Studies on yeast TPE indicate that the repressive properties of such a newly formed telomere are shared by native telomeres, except for chromosome-specific modulations exerted by the natural subtelomeric regions (Fourel et al., 1999; Pryde and Louis, 1999). Therefore, hTPE is highly likely to be a general property of human telomeres.
A specific chromatin structure determines hTPE
Trichostatin A (TSA) inhibits the HDAC families of histone deacetylases and leads to the loss of structure of the heterochromatin compartment (Yoshida et al., 1990; Taddei et al., 2001). An increase of EGFP expression rapidly occurs in a population of 4C11 cells after a first treatment with a subtoxic dose of TSA (Figure 5). Impressively, the derepression induced by TSA is exacerbated upon a second treatment (Figure 5), indicating a potentiation of the effect. No such phenomenon was detected in populations of 2A3 or 3B10 cells (Figure 5). This demonstrates that it is not an intrinsic property of the EGFP reporter cassette to be highly sensitive to TSA and indicates that the TSA effect in 4C11 cells is dependent on the telomeric location of the EGFP gene. A similar conclusion was reported in HeLa cells (Baur et al., 2001). In contrast, no apparent change in EGFP expression was observed when cells were treated by two or more successive doses of 5 azacytidin (5 azaC), which reduces cytosine methylation (Harrison et al., 1983), and of sirtinol, a drug recently identified as a specific inhibitor of the sir2/sirtuin family of NAD-dependant deacetylases (Grozinger et al., 2001).
Figure 5.

TSA alleviates telomeric silencing. Populations of 4C11, 2A3 or 3B10 cells sorted for a deprivation of EGFP-positive cells were incubated for 3 days with a culture medium containing the indicated drug at T1; then the medium was changed with one of identical composition (T2).
In order to monitor the impact of the TSA treatments on the spatial organization of the centromeres and telomeres in our cellular system, FISH experiments with alphoid DNA and telomeric PNA probes were performed. Remarkably, each of the three types of treatments correlates with distinct patterns of heterochromatin destructuration (see Supplementary figure 2A). These changes occur at the first TSA treatment and do not seem to be exacerbated at the second treatment. The spotty pattern of telomeric DNA appears unchanged upon the two successive TSA treatments (see Supplementary figure 2B). These results prompted us to determine the localization of the three mammalian isoforms of the heterochromatin protein 1 (HP1α, β and γ). HP1α and HP1β localize predominantly to centromeric heterochromatin, and HP1γ localizes to both heterochromatin and euchromatin (Minc et al., 1999; Aagard et al., 2000). Importantly, they bind heterochromatin in a TSAsensitive way (Taddei et al., 2001). In interphase nuclei, the three HP1 isoforms exhibit a punctated pattern, whereas, in metaphase chromosomes, HP1α and HP1β show a predominant centromeric staining (see Supplementary figure 2C). As previously reported (Minc et al., 1999; Aagard et al., 2000), we also noted infrequent signals at chromosome tips. This observation indicates that HP1α and HP1β play a role at telomeres.
In contrast to alphoid DNA, the first treatment with TSA does not seem to greatly alter the HP1 pattern in interphase nuclei. However, the second TSA treatment clearly leads to a release of HP1α and HP1β but not of HP1γ (see Supplementary figure 2C) We conclude that the first treatment of TSA is sufficient to perturb the organization of centromeric DNA but is insufficient to massively dissociate HP1α and HP1β from centromeres. Interestingly, the correlation between hTPE potentiation and HP1α/β release at the time of the second TSA treatment indicates an involvement of HP1 isoforms in hTPE.
Finally, we investigated whether hTPE can be related to a preferential association of the reporter gene with the centromeric heterochromatin compartment. The proportion of subtelomeric EGFP genes associated with various types of centromeric regions remains essentially unchanged in EGFP-positive and -negative cells or when cells are treated with TSA (see Supplementary figure 3). Thus, there is no apparent correlation between the silencing state of the subtelomeric gene and its association with centromeric heterochromatin. This does not favor a model where hTPE would result from the recruitment of chromosome ends at or near centromeres (Brown et al., 1997). However, it is plausible that the silent telomere localizes to alternative areas of heterochromatin. In particular, the telomere itself might recruit one or several repressive factors in a TSAsensitive way. The correlation between the HP1 delocalization and hTPE alleviation, together with the occasional staining of HP1 isoforms to telomeres, indicate that one of these factors could be HP1.
Methods
Plasmids and molecular biology.
The pCMV and pCMVTelo plasmids derived from pEGFP1 (Clontech) (Figure 1A) carry the HyTK gene excised from pNYH3 (Lupton et al., 1991) (obtained through the courtesy of T. de Lange). The pITRF1 results from the cloning of a FLAG version of the TERF1 gene (Vassetzky et al., 1999) into pIRES1neo (Clontech). Details concerning the exact cloning procedure will be given upon request. Cellular DNA was prepared by using Nucleon Bacc2 kits (Amersham). FISH experiments were performed with biotin or digoxigenin labeled pCMV plasmids or with a chromosomespecific set of probes (Vysis).
Cells methods.
The human cell line C33-A (carcinoma, cervix) was grown in DMEM with bicarbonate supplemented with 10% fetal calf serum, 1% L- glutamine and antibiotics. Transfections were performed by using a modified calcium phosphate method (Chen and Okayama, 1987). The transfection efficiency of C33-A cells was estimated by transfecting 2 μg of a LacZ-expressing vector and was found to vary between 30 and 40% in independent experiments. The CAT expression was determined by ELISA (Boehringer). The percentage of EGFP-positive cells was monitored by Facscan. Populations of cells either enriched or deprived in EGFP-positive cells were sorted through Facstar +.
To determine the highest subtoxic concentration of each drug (TSA, 5 azaC and sirtinol) that was non-toxic and did not affect cell growth, a C33-A cell line was cultured in various concentrations of the drug for 1 week. The highest concentrations that did not cause a decrease in cell growth rate were chosen.
Supplementary Material
Supplementary figure 1
Acknowledgments
We would like to thank S. Gasser for a critical reading of the manuscript; T. de Lange for pNYH3; A. Sergeant for pBLCat; E. Viegas-Pequignot and F. Palladino for HP1 antibodies; and A. Thomas, S. Mouradian and P. Barba for cytometry facilities. The work in E.G.'s laboratory was supported by La Ligue Nationale contre le Cancer and the Association Française contre les Myopathies (AFM). S.B. is supported by a postdoc fellowship from Centre L. Bérard. The work in J.F.P.'s laboratory was supported by Cofin 98-MURST and Programma Biomolecole per la Salute Umana. M.P.Z. was in part supported by a MURST 'Galileo' doctoral fellowship. The work in L.S.'s laboratory is supported by CEC funding: FIGH-CT-1999-0009.
References
- Aagaard L., Schmid M., Warburton P. and Jenuwein T. (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci., 113, 817–829. [DOI] [PubMed] [Google Scholar]
- Baur J.A., Zou Y., Shay J.W. and Wright W.E. (2001) Telomere position effect in human cells. Science, 292, 2075–2077. [DOI] [PubMed] [Google Scholar]
- Bayne R.A., Broccoli D., Taggart M.H., Thomson E.J., Farr C.J. and Cooke H.J. (1994) Sandwiching of a gene within 12 kb of a functional telomere and α satellite does not result in silencing. Hum. Mol. Genet., 3, 539–546. [DOI] [PubMed] [Google Scholar]
- Bilaud T., Koering C.E., Binet-Brasselet E., Ancelin K., Pollice A., Gasser S.M. and Gilson E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H. (2000) The end of the (DNA) line. Nat. Struct. Biol., 7, 847–850. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M. and Fisher A.G. (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell, 91, 845–854. [DOI] [PubMed] [Google Scholar]
- Chen C. and Okayama H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol., 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P. and de Lange T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cryderman D.E., Morris E.J., Biessmann H., Elgin S.C. and Wallrath L.L. (1999) Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J., 18, 3724–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr C., Fantes J., Goodfellow P. and Cooke H. (1991) Functional reintroduction of human telomeres into mammalian cells. Proc. Natl Acad. Sci. USA, 88, 7006–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.M. and Fangman W.L. (1992) A position effect on the time of replication origin activation in yeast. Cell, 68, 333–339. [DOI] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C.E. and Gilson E. (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J., 18, 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D., Green M.R. and Tupler R. (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscles. Cell, 110, 339–348. [DOI] [PubMed] [Google Scholar]
- Gilson E., Laroche T. and Gasser S. (1993) Telomeres and the functional architecture of the nucleus. Trends Cell Biol., 3, 128–134. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L. and Zakian V.A. (1990) Position effect at S. cerevisiae telomeres: reversible represssion of Pol II transcription. Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- Grozinger C.M., Chao E.D., Blackwell H.E., Moazed D. and Schreiber S.L. (2001) Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem., 276, 38837–38843. [DOI] [PubMed] [Google Scholar]
- Hanish J.P., Yanowitz J.L. and de Lange T. (1994) Stringent sequence requirements for the formation of human telomeres. Proc. Natl Acad. Sci. USA, 91, 8861–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.J., Anisowicz A., Gadi I.K., Raffeld M. and Sager R. (1983) Azacytidine-induced tumorigenesis of CHEF/18 cells: correlated DNA methylation and chromosome changes. Proc. Natl Acad. Sci. USA, 80, 6606–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res., 451, 227–240. [DOI] [PubMed] [Google Scholar]
- Lupton S.D., Brunton L.L., Kalberg V.A. and Overell R.W. (1991) Dominant positive and negative selection using a hygromycin phosphotransferase–thymidine kinase fusion gene. Mol. Cell. Biol., 11, 3374–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E., Allory Y., Worman H.J., Courvalin J.C. and Buendia B. (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma, 108, 220–234. [DOI] [PubMed] [Google Scholar]
- Nimmo E.R.N., Cranston G. and Allshire R.C. (1994) Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J., 13, 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir R., Wong A.C., McDermid H.E., Skorecki K.L. and Selig S. (1999) Position effect of human telomeric repeats on replication timing. Proc. Natl Acad. Sci. USA, 96, 11434–11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F.E. and Louis E.J. (1999) Limitations of silencing at native yeast telomeres. EMBO J., 18, 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G., Blundell P.A., Dirks-Mulder A., Kieft R. and Borst P. (1995) A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell, 83, 547–553. [DOI] [PubMed] [Google Scholar]
- Smith Z.E. and Higgs D.R. (1999) The pattern of replication at a human telomeric region (16p13.3): its relationship to chromosome structure and gene expression. Hum. Mol. Genet., 8, 1373–1386. [DOI] [PubMed] [Google Scholar]
- Sprung C.N., Sabatier L. and Murnane J.P. (1996) Effect of telomere length on telomeric gene expression. Nucleic Acids Res., 24, 4336–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Maison C., Roche D. and Almouzni G. (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol., 3, 114–120. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- Vassetzky N.S., Gaden F., Brun C., Gasser S.M. and Gilson E. (1999) Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res., 27, 4687–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L., Perricaudet M., Sergeant A. and Manet E. (1996) Epstein–Barr virus EBNA3A and EBNA3C proteins both repress RBP-J κ-EBNA2-activated transcription by inhibiting the binding of RBP-J κ to DNA. J. Virol., 70, 5909–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W.E., Tesmer V.M., Liao M.L. and Shay J.W. (1999) Normal human telomeres are not late replicating. Exp. Cell Res., 251, 492–499. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Akita M. and Beppu T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem., 265, 17174–17179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1


