Abstract
In mammals, growth-dependent regulation of rRNA synthesis is brought about by the transcription initiation factor TIF-IA. TIF-IA is associated with a fraction of the TBP-containing factor TIF-IB/SL1 and the initiation-competent form of RNA polymerase I (Pol I). We investigated the mechanisms that down-regulate cellular pre-rRNA synthesis and demonstrate that nutrient starvation, density arrest and protein synthesis inhibitors inactivate TIF-IA and impair the association of TIF-IA with Pol I. Moreover, we used a panel of TIF-IA deletion mutants to map the domains that mediate the interaction of TIF-IA with Pol I and TIF-IB/SL1. We found that amino acids 512–609 interact with two subunits of Pol I, RPA43 and PAF67, whereas a short, conserved motif (LARAK, amino acids 411–415) is required for the association of TIF-IA with TAFI95 and TAFI68. The results uncover an interphase for essential protein–protein interactions that facilitate Pol I preinitiation complex formation.
Introduction
Preinitiation complex formation at the mammalian ribosomal RNA gene promoter is nucleated by the synergistic action of two DNA binding proteins, the HMG-box-containing upstream binding factor UBF (Jantzen et al., 1992) and the RNA polymerase I (Pol I)specific TBP–TAFI complex TIF-IB/SL1 (Comai et al., 1992; Eberhard et al., 1993). Pol I, together with two associated initiation factors, TIF-IA and TIF-IC, is recruited to the transcription start site by specific interaction with UBF and TIF-IB/SL1 bound to the core element of the rDNA promoter (for a review, see Grummt, 1999). TIF-IA was initially characterized as an activity that complements inactive extracts from quiescent mouse cells (Buttgereit et al., 1985). Extracts prepared from serumstarved or cycloheximide-treated cells lack this activity and are therefore transcriptionally inactive. TIF-IA corresponds to factor C* (Brun et al., 1994) and yeast Rrn3p (Yamamoto et al., 1996). Both in yeast and mammals, Rrn3p/TIF-IA associates with a subpopulation of Pol I to form the transcriptionally active enzyme, defined as the Pol I entity that is capable of initiating transcription from the rDNA promoter.
Besides being associated with Pol I, TIF-IA has been shown to interact with TIF-IB/SL1 (Miller et al., 2001). This suggests that, by interacting with both Pol I and TIF-IB/SL1, TIF-IA targets transcriptionally active Pol I molecules to the rDNA promoter. Given the essential role of TIF-IA in adapting rDNA transcription to cell growth, we undertook a detailed analysis of the protein domains that mediate the interaction of TIF-IA with Pol I and TIF-IB/SL1. We demonstrate that the C-terminal part of TIF-IA interacts with two subunits of Pol I, RPA43 and PAF67, whereas an internal region of TIF-IA (LARAK, amino acids 411–415) mediates the interaction with TAFI68 and TAFI95. Importantly, the interaction between TIF-IA and Pol I is impaired in stationary, starved and cycloheximide-treated cells, underscoring the biological significance of these protein–protein interactions in initiation complex formation.
Results and Discussion
TIF-IA activity is down-regulated in growth-arrested cells
Fluctuations in cellular rRNA synthesis have been observed under a variety of conditions that affect cell growth and metabolism. Previous studies revealed that the shutdown of rRNA synthesis during mitosis and early G1 phase is due to inactivation of TIF-IB/SL1 and UBF (Heix et al., 1998; Klein and Grummt, 1999). Growth-dependent regulation of Pol I, on the other hand, has been attributed to alterations in the amount or activity of TIF-IA (Buttgereit et al., 1985; Schnapp et al., 1993). To study the molecular mechanisms mediating growth-dependent regulation of Pol I transcription, pre-rRNA levels of exponentially growing NIH 3T3 cells were compared with those of serumstarved, cycloheximide-treated and density-arrested cells. Pre-rRNA synthesis was monitored on northern blots using a labeled probe that hybridizes to the 5′ end of 45S pre-rRNA and specifically detects unprocessed pre-rRNA molecules. Figure 1A shows that pre-rRNA synthesis was markedly decreased in serumstarved, cycloheximide-treated and density-arrested cells. This reduction of rRNA synthetic activity supports early studies demonstrating that conditions that harm cellular metabolism impair Pol I transcription (Yu and Feigelson, 1972).
Figure 1.
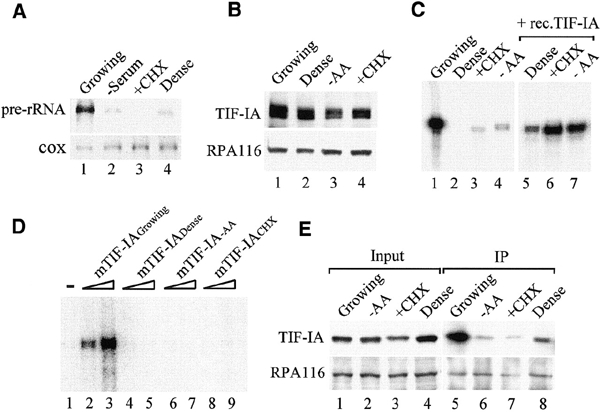
TIF-IA is inactivated in growth-arrested cells. (A) Northern blot. 45S pre-rRNA levels were monitored in 15 μg of RNA from exponentially growing, serum-starved, cycloheximide-treated and density-arrested cells. To normalize for variations of RNA loading, the blot was also hybridized with a probe complementary to cytochrome C oxidase (cox) mRNA. (B) Western blot. TIF-IA and Pol I from exponentially growing (lane 1), density-arrested (lane 2), amino-acid-starved (lane 3) or cycloheximide-treated (lane 4) FM3A cells were visualized on immunoblots with α-TIF-IA or α-RPA116 antibodies. (C) In vitro transcription. Nuclear extract proteins (50 μg) from growing (lane 1), density-arrested (lanes 2 and 5), cycloheximide-treated (lanes 3 and 6) and amino-acid-starved (lanes 4 and 7) FM3A cells were assayed for transcriptional activity in the absence (lanes 1–4) or presence (lanes 5–7) of 30 ng of recombinant TIF-IA. (D) TIF-IA is targeted by extracellular signals that inhibit growth. TIF-IA (30 and 60 ng) purified from exponentially growing (lanes 2 and 3), density-arrested (lanes 4 and 5), amino-acid-starved (lanes 6 and 7) and cycloheximide-treated (lanes 8 and 9) NIH 3T3 cells were assayed for their capability to restore the transcriptional activity of a nuclear extract from density-arrested FM3A cells (lane 1). (E) The interaction between TIF-IA and Pol I is impaired in growth-arrested cells. NIH 3T3 cells (1 × 106) were transfected with 2 μg of pcDNA3.1-Flag-hTIF-IA. To achieve confluency, 8 × 106 cells were seeded onto a 100 mm plate 12 h after transfection. Alternatively, cells were treated with cycloheximide or starved of cystine and methionine for 2 h before harvesting. Pol I was immunoprecipitated from cell lysates with α-RPA53 antibodies, and TIF-IA and Pol I were detected on western blots.
To investigate whether the amount or activity of TIF-IA was altered in response to changes in cell growth, TIF-IA levels and transcriptional activity were compared in nuclear extracts from density-arrested, amino-acid-starved or cycloheximide-treated cells. All four extracts contained similar levels of TIF-IA and Pol I (Figure 1B) but exhibited marked differences in transcriptional activity. Extracts from growing cells efficiently transcribed rDNA (Figure 1C, lane 1), whereas extracts from dense, cycloheximide-treated or starved cells were virtually inactive (lanes 2–4). Significantly, the addition of recombinant TIF-IA restored transcriptional activity, reaching levels that are comparable to the control extract (lanes 5–7). This result demonstrates that TIF-IA, but none of the other components of the Pol I transcription machinery, is targeted by diverse signaling pathways that ultimately down-regulate the cell's biosynthetic activity.
If this conclusion is correct, then TIF-IA from growth-arrested cells should be transcriptionally inactive. To test this, TIF-IA was immunopurified from growing, confluent, starved and cycloheximide-treated NIH 3T3 cells, and equal amounts of TIF-IA were assayed for their capability to restore Pol I transcription in extracts from stationary cells. TIF-IA from growing cells activated Pol I transcription (Figure 1D, lanes 2 and 3). In contrast, TIF-IA from density-arrested, starved and cycloheximide-treated cells was transcriptionally inactive (lanes 4–9). Thus, the activity, but not the amount, of TIF-IA is regulated by signals that impair cell metabolism and growth.
TIF-IA has been shown to be part of the Pol I 'holoenzyme', the enzyme moiety that is associated with most, if not all, factors required for transcription initiation (Saez-Vasquez and Pikaard, 1997; Seither et al., 1998; Albert et al., 1999). Moreover, TIF-IA has been shown to interact with TIF-IB/SL1 (Miller et al., 2001), which suggests that TIF-IA bridges both protein complexes. To assess the function of TIF-IA in initiation complex formation, Pol I was precipitated from exponentially growing, confluent, starved or cycloheximide-treated cells, and coprecipitated TIF-IA was detected on western blots. Consistent with TIF-IA being associated with Pol I, significant amounts of TIF-IA were present in the immunoprecipitates from growing cells (Figure 1E, lane 5). Importantly, the amount of TIF-IA that was associated with Pol I markedly decreased in growth-arrested cells (lanes 6–8). This result is consistent with recent data in yeast and mammals demonstrating dissociation of TIF-IA from Pol I in stationary or cycloheximide-treated cells (Milkereit and Tschochner, 1998; Cavanaugh et al., 2002). The current view is that inhibition of cell growth leads to hypophosphorylation and inactivation of TIF-IA and impairs the association of TIF-IA with Pol I.
TIF-IA interacts with two subunits of Pol I
To study the interaction of TIF-IA with Pol I and TIF-IB/SL1, Flag-tagged TIF-IA was incubated with cellular fractions containing partially purified Pol I or TIF-IB/SL1, precipitated with anti-FLAG antibodies, and coprecipitated Pol I and TIF-IB/SL1 were analyzed on western blots. Consistent with TIF-IA mediating the interaction between Pol I and TIF-IB/SL1, both Pol I and TIF-IB/SL1 coprecipitated with TIF-IA (Figure 2A).
Figure 2.
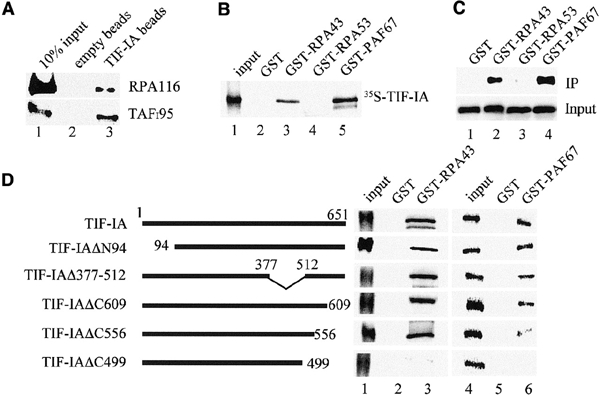
The C-terminus of TIF-IA interacts with RPA43 and PAF67. (A) Interaction of TIF-IA with Pol I and TIF-IB/SL1. Recombinant TIF-IA was incubated with 50 μl of partially purified cellular Pol I or TIF-IB/SL1 and incubated with control beads (lane 2) or bead-bound α-TIF-IA antibodies (lane 3). Associated Pol I and TIF-IB were visualized on western blots using α-RPA116 or α-TAFI95 antibodies. (B) GST pull-down assays. The indicated GST fusion proteins were immobilized on glutathione–Sepharose and incubated with 35S-labeled TIF-IA. TIF-IA bound to GST (lane 2), GST–RPA43 (lane 3), GST–RPA53 (lane 4) and GST–PAF67 (lane 5) was visualized by autoradiography. (C) RPA43 and PAF67 form a stable complex with TIF-IA. Extracts from E. coli expressing TIF-IA and GST (lane 1), TIF-IA and GST–RPA43 (lane 2), TIF-IA and GST–RPA53 (lane 3) or TIF-IA and GST–PAF67 (lane 4) were assayed on western blots for TIF-IA levels (lower panel). After incubation with glutathione–Sepharose, associated TIF-IA was monitored on western blots (upper panel). (D) Mutational analysis of TIF-IA/Pol I interactions. 35S-labeled TIF-IA and the respective mutants were incubated with immobilized GST (lanes 2 and 5), GST–RPA43 (lane 3) and GST–PAF67 (lane 6). TIF-IA was analyzed by SDS–PAGE and autoradiography. Twenty percent of the input proteins are shown in lanes 1 and 4.
In Saccharomyces cerevisiae, Rrn3p has been found to interact with RPA43, a unique subunit of Pol I (Peyroche et al., 2000; Fath et al., 2001). Given that functionally important protein–protein interactions are evolutionarily conserved, we examined whether the homologous mammalian subunit mediates the interaction with TIF-IA. To monitor interactions between TIF-IA and subunits of Pol I, pull-down experiments were performed (Figure 2B). 35S-labeled TIF-IA interacted with GST–mRPA43 but not with GST (lanes 2 and 3). No interaction was observed with RPA53, the third largest subunit of Pol I (lane 4). Surprisingly, the strongest binding was observed to PAF67 (lane 5), a 67 kDa protein that is tightly associated with the initiation-competent form of Pol I and was suggested to play an essential role in recruiting Pol I to the rDNA promoter (Seither et al., 2001).
The interaction of TIF-IA with both PAF67 and RPA43 was substantiated by co-expressing TIF-IA in Escherichia coli together with GST fusion proteins encoding PAF67, RPA53 or mRPA43 and capturing bound TIF-IA on glutathione–Sepharose. Consistent with the results above, TIF-IA was bound to GST–PAF67 and GST–RPA43 but not to control beads or GST–RPA53 (Figure 2C). Again, TIF-IA bound more efficiently to GST–PAF67 than to GST–RPA43. Thus, TIF-IA interacts with two polypeptides of Pol I, e.g. RPA43, a genuine subunit and PAF67, a Pol I-associated factor that marks the initiation-competent enzyme moiety. This suggests that, by interacting with PAF67, TIF-IA may target a functional subset of Pol I molecules into the initiation complex.
To map the respective domains of TIF-IA that interact with RPA43 and PAF67, GST pull-down experiments were performed with a set of TIF-IA deletion mutants. As shown in Figure 2D, mutants lacking the N-terminus (TIF-IA/ΔN94), a large internal region (TIF-IA/Δ377–512) or the C-terminal region (TIF-IA/ΔC609) efficiently associate with both RPA43 and PAF67. Deletion of 95 amino acids from the C-terminus (TIF-IA/ΔC556) reduces binding to PAF67 without affecting the interaction with RPA43. Further truncation of the C-terminus (TIF-IA/ΔC499) abolishes binding of TIF-IA to both RPA43 and PAF67. This suggests that two domains located between residues 512 and 609 mediate the interaction between TIF-IA and Pol I.
A conserved sequence element (LARAK) mediates the interaction between TIF-IA and TIF-IB
To examine the interaction of TIF-IA with subunits of TIF-IB, immobilized TIF-IA was incubated with extracts from Sf9 cells that overexpress either of the four subunits of TIF-IB. Bound proteins were analyzed on western blots using antibodies against TAFI95, TAFI68, TAFI48 and TBP, respectively. As demonstrated in Figure 3A, TIF-IA interacted with TAFI95 and TAFI68, whereas no interaction of TIF-IA with TAFI48 or TBP was observed.
Figure 3.
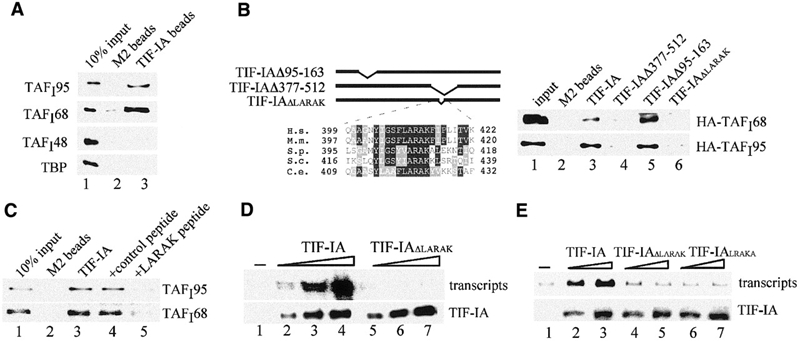
A conserved motif (LARAK) mediates the interaction of TIF-IA with TAFI95 and TAFI68. (A) Pull-down assay. Bead-bound Flag-tagged TIF-IA was incubated with extracts from Sf9 cells overexpressing HA-tagged TAFI48, TAFI68, TAFI95 or TBP, respectively. Proteins were separated by SDS–PAGE, and TAFIs were visualized on western blots using α-HA antibodies (lane 3). M2 beads saturated with the Flag epitope peptide were used as a control (lane 2). (B) The LARAK motif mediates the interaction of TIF-IA with TAFI68 and TAFI95. Flag-tagged TIF-IA (lane 3), TIF-IAΔ377–512 (lane 4), TIF-IAΔ95–163 (lane 5) and TIF-IAΔLARAK (lane 6) were incubated with HA-tagged TAFI68 or TAFI95. TIF-IA was immunoprecipitated, and associated TAFIs were visualized on western blots. A scheme of the mutants and a sequence alignment of the LARAK domain of TIF-IA homologues are shown above. H.s., Homo sapiens; M.m., Mus musculus; S.p., Schizosaccharomyces pombe; S.c., Saccharomyces cerevisiae; C.e., Caenorhabditis elegans. (C) A synthetic peptide (LARAK) blocks the interaction between TIF-IA and TIF-IB/SL1. Immobilized TIF-IA was incubated at 4°C for 4 h with partially purified TIF-IB/SL1 in the absence (lane 3) or presence of 200 ng control peptide (EHLWKKLQDPSNPAI, lane 4) or LARAK peptide (YIGSFLARAKFITVKSC, lane 5) in 250 μl of buffer AM-100/0.5% NP-40. After stringent washing, bound TIF-IB/SL1 was analyzed on immunoblots with α-TAFI95 or α-TAFI68 antibodies. (D) The LARAK motif is essential for TIF-IA activity. In vitro transcription assays contained nuclear extracts from density-arrested FM3A cells and no exogenous TIF-IA (lane 1) or 15, 30 and 50 ng of wild-type TIF-IA (lanes 2–4) or TIF-IAΔLARAK (lanes 5–7). The amounts of recombinant TIF-IA added to the reactions was monitored on western blots with α-Flag antibodies (lower panel). (E) Deletion or permutation of the LARAK motif abrogates TIF-IA activity in vivo. NIH 3T3 cells were cotransfected with 2.5 μg of the rDNA reporter plasmid and 1 or 2 μg of pBK-CMV-Flag-TIF-IA (lanes 2 and 3), pBK-CMV-Flag-TIF-IAΔLARAK (lanes 4 and 5) or pBK-CMV-Flag-TIF-IALRAKA (lanes 6 and 7). Transcripts from the rDNA reporter were monitored on northern blots. TIF-IA expression levels were monitored on western blots with α-Flag antibodies (lower panel).
To define the domains of TIF-IA that mediate the interaction with TAFIs, wild-type and mutant TIF-IA were expressed in Sf9 cells and incubated with cell lysates containing HA-tagged TAFI95 or TAFI68, and bound TAFIs were monitored on immunoblots. Truncation of the N- or C-terminal part of TIF-IA did not affect the interaction with either subunit (data not shown). Similarly, a mutant lacking amino acids 95–163 bound to both subunits with an efficiency comparable with wild-type TIF-IA (Figure 3B, lane 5). In contrast, deletion of residues 377–512 abolished binding (lane 4). A search for conserved sequence elements within amino acids 377–512 revealed a motif (F/Y(L)ARAK) that is highly conserved among different species (Figure 3B). Deletion of the LARAK sequence (amino acids 411–415) abrogated TIF-IA binding to both TAFIs (Figure 3B, lane 6), indicating that this motif serves an essential role in preinitiation complex assembly.
Given that the LARAK motif mediates the interaction between TIF-IA and TIF-IB/SL1, a synthetic peptide harboring the LARAK sequence should prevent the interaction of both factors. Indeed, the LARAK peptide blocked the association of TIF-IA with TIF-IB/SL1 (Figure 3C, lane 5), whereas a control peptide had no effect (lane 4). Thus, amino acids 411–415 are required for the association of TIF-IA with TIF-IB.
Consistent with the functional significance of the LARAK motif, TIF-IAΔLARAK was transcriptionally inactive. TIF-IAΔLARAK was incapable of complementing inactive extracts from density-arrested cells in vitro (Figure 3D). Moreover, when assayed for activation of a Pol I reporter gene in vivo, wild-type but not TIF-IAΔLARAK stimulated Pol I transcription (Figure 3E, lanes 1–5). Converting the LARAK sequence to LRAKA, a permutation that does not alter the charge of TIF-IA, also abolished TIF-IA activity (lanes 6 and 7). These results demonstrate that the interaction between TIF-IA and TIF-IB is mediated by a discrete number of amino acids that have been evolutionary conserved.
The finding that both TIF-IA and the yeast homologue Rrn3 is a key player in recruiting Pol I to the rDNA promoter underscores the essential role of this transcription factor in rRNA synthesis. TIF-IA activity is regulated by diverse extracellular signals, indicating that this factor adapts Pol I transcription to cell growth. TIF-IA is phosphorylated at multiple sites (unpublished data), and data in yeast have revealed that reversible phosphorylation of both Rrn3 and Pol I mediate formation and dissociation of the initiation complex (Fath et al., 2001). Therefore, elucidating the chains of events by which extracellular signals are transferred into the nucleolus and modify the activity of Rrn3/TIF-IA will unravel the mechanism of Pol I transcription initiation and the pathways that control this essential process.
Methods
Plasmids.
The cDNA encoding TIF-IA (DDB/EMBL/GenBank database accession No. AJ272050) was tagged with the Flag-epitope and subcloned into various expression vectors. Mutant forms of TIF-IA were generated by in vitro mutagenesis using standard techniques. The rDNA reporter pMr1930-BH has been described previously (Budde and Grummt, 1999). The cDNA encoding amino acids 1–248 of mouse RPA43 was amplified from plasmid IMAGp998M106538 (RZPD).
Cell culture and transfection experiments.
HEK 293T or NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM glutamine. Cells were harvested either in the log-phase or after reaching confluence. Alternatively, cells were starved of cystine and methionine or treated with 0.1 mg/ml cycloheximide for 2 h. For in vivo analysis of wild-type and mutant TIF-IA, 5 × 105 cells were cotransfected with 2.5 μg of reporter plasmid (pMr1930-BH) and different amounts of a CMV-based expression vector encoding tagged wild-type or mutant forms of TIF-IA. After 48 h, cellular RNA was isolated and hybridized to 32P-labeled probes that are complementary to pUC sequences inserted between the rDNA promoter and terminator in pMr1930-BH.
Expression and purification of recombinant TIF-IA.
The expression and purification of Flag-tagged TIF-IA from insect cells has been described previously (Bodem et al., 2000). To immunopurify cellular TIF-IA, 5 × 106 NIH 3T3 cells were lysed for 45 min at 4°C in 0.8 ml of 20 mM Tris–HCl pH 7.4, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100 in the presence of protease inhibitors. After centrifugation for 30 min at 10 000 g, the supernatants were incubated for 4 h with α-TIF-IA antibodies (10 μg) bound to 20 μl of protein G-agarose. After washing, the beads were resuspended in 200 μl of buffer AM-100 and assayed for transcriptional activity.
In vitro transcription assays.
Standard reactions (25 μl) contained 30 ng of template pMr600/EcoRI, 12 mM Tris–HCl pH7.9, 0.1 mM EDTA, 0.5 mM dithioerythritol, 5 mM MgCl2, 80 mM KCl, 12% glycerol, 0.66 mM each of ATP, CTP and GTP, 0.01mM UTP, 1 μCi [α-32P]UTP and 50 μg of nuclear extract proteins from FM3A cells.
Immunoprecipitation and immunoblotting.
Immunoprecipitations were carried out using nuclear extracts from FM3A cells, lysates from NIH 3T3 cells, or HEK 293T cells overexpressing Flag-tagged TIF-IA. Cells were lysed in 1 ml of immunoprecipitation buffer (50 mM Tris-HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, 1 mM DTT) and incubated for 4 h at 4°C with the respective antibodies. After extensive washing, precipitated proteins were analyzed on immunoblots.
Protein–protein interaction experiments.
Immobilized Flag-tagged TIF-IA and control beads saturated with the Flag-epitope peptide were incubated with lysates from Sf9 cells overexpressing recombinant subunits of TIF-IB. After incubation for 2 h at 4°C in buffer AM-100 plus 0.5% NP40 and washes, associated proteins were identified on western blots. To examine the association of TIF-IA with Pol I and TIF-IB, 500 ng of TIF-IA expressed in Sf9 cells were incubated for 2 h at 4°C with 50 μl of partially purified Pol I or TIF-IB (H-400 or CM-400 fraction; Schnapp and Grummt, 1996). After precipitation with anti-Flag antibodies, associated proteins were identified on western blots. To monitor the interaction between TIF-IA and subunits of Pol I in vitro, GST, GST–RPA43, GST–RPA53 and GST–PAF67 were attached to glutathione–Sepharose, and 10 μl of packed beads containing 5 μg of GST or GST fusion proteins were incubated with 7 μl of 35S-labeled TIF-IA in 100 μl of buffer (50 mM Tris–HCl pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitors). After incubation for 1 h at room temperature, beads were washed, and bound proteins were analyzed by SDS–PAGE and autoradiography. Alternatively, TIF-IA and GST fusion proteins were co-expressed in E. coli and bound to glutathione–Sepharose as described previously (Peyroche et al., 2000).
Acknowledgments
We thank Bettina Dörr for preparation and fractionation of cell extracts. We are grateful to all members of the laboratory for sharing reagents and advice. This work was supported by the German Cancer Research Center, the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
References
- Albert A.C., Denton M., Kermekchiev M. and Pikaard C.S. (1999) Histone acetyltransferase and protein kinase activities copurify with a putative Xenopus RNA polymerase I holoenzyme selfsufficient for promoter-dependent transcription. Mol. Cell. Biol., 19, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem J., Dobreva G., Hoffmann-Rohrer U., Iben S., Zentgraf H., Delius H., Vingron M. and Grummt I. (2000) TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO rep., 1, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R.P., Ryan K. and Sollner-Webb B. (1994) Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol., 14, 5010–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A. and Grummt I. (1999) p53 represses ribosomal gene transcription. Oncogene, 18, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Buttgereit D., Pflugfelder G. and Grummt I. (1985) Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res., 13, 8165–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh A.H., Hirschler-Laszkiewicz I., Hu Q., Dundr M., Smink T., Misteli T. and Rothblum L.I. (2002) Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem., 277, 27423–27432. [DOI] [PubMed] [Google Scholar]
- Comai L., Tanese N. and Tjian R. (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor SL1. Cell, 68, 965–976. [DOI] [PubMed] [Google Scholar]
- Eberhard D., Tora L., Egly J.M. and Grummt I. (1993) A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res., 21, 4180–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S., Milkereit P., Peyroche G., Riva M., Carles C. and Tschochner H. (2001) Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl Acad. Sci. USA, 98, 14334–14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. (1999) Regulation of mammalian ribosomal gene transcription. Prog. Nucleic Acid Res. Mol. Biol., 62, 109–154. [DOI] [PubMed] [Google Scholar]
- Heix J., Vente R., Voit R., Budde A., Michaelidis T.M. and Grummt I. (1998) Mitotic silencing of rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 17, 7373–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H.M., Chow A.M., King D.S. and Tjian R. (1992) Multiple domains of the RNA polymerase I activator hUBF interacts with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6, 1950–1963. [DOI] [PubMed] [Google Scholar]
- Klein J. and Grummt I. (1999) Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl Acad. Sci. USA, 96, 6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner H. (1998) A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J., 17, 3692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Panov K.I., Friedrich J.K., Trinkle-Mulcahy L., Lamond A.I. and Zomerdijk J.C. (2001) hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G., Milkereit P., Bischler N., Tschochner H., Schultz P., Sentenac A., Carles C. and Riva M. (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J., 19, 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J. and Pikaard C.S. (1997) Extensive purification of putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl Acad. Sci. USA, 94, 11869–11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A. and Grummt I. (1996) Purification, assay, and properties of RNA polymerase I and class Ispecific transcription factors in mouse. Methods Enzymol., 273, 233–248. [DOI] [PubMed] [Google Scholar]
- Schnapp A., Schnapp G., Erny B. and Grummt I. (1993) Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol., 13, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seither P., Iben S. and Grummt I. (1998) Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol., 275, 43–53. [DOI] [PubMed] [Google Scholar]
- Seither P., Iben S., Thiry M. and Grummt I. (2001) PAF67, a novel protein that is associated with the initiation-competent form of RNA poly-merase I. Biol. Chem., 382, 1163–1170. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi Y., Dodd J.A. and Nomura M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Yu F.L. and Feigelson P. (1972) The rapid turnover of RNA polymerase of rat liver nucleolus and of its messenger RNA. Proc. Natl Acad. Sci. USA, 69, 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]


