Abstract
Mitotic and meiotic inheritance of epigenetic information is coupled to the reproduction of chromatin conformation and DNA methylation patterns. This implies that the S phase of the cell cycle provides a window of opportunity for changes in epigenetic determination. Recent studies, however, have suggested that chromatin structure is also rather dynamic in quiescent cells of multicellular eukaryotes and that silent heterochromatic regions can become accessible to transcription. Such epigenetic flexibility in differentiated tissues could be of physiological importance. The mechanisms and molecular components involved are of great interest but as yet unknown. We examined MOM1 (Morpheus' Molecule 1), a regulator of transcriptional gene silencing (TGS) that acts independently of DNA methylation, for its role in the maintenance of TGS in non-dividing, differentiated cells. The results provide evidence that TGS maintenance mediated by MOM1 is a dynamic process that can be modified in non-dividing cells of mature plant organs by depletion of MOM1.
Introduction
Epigenetic inheritance exploits the reproduction of a particular chromatin conformation during its assembly on the newly replicated DNA (for a review, see Grewal and Elgin, 2002). In mammals and plants, post-replicative reproduction of chromatin structure seems to be linked to DNA methylation patterns copied by DNA methyltransferases present in DNA replication foci (Kass et al., 1997; Araujo et al., 1998). Although rapid, hormone-induced changes of chromatin accessibility suggest its dynamic nature (for a review, see Xu et al., 1999), mitotically and possibly meiotically heritable modification of global epigenetic patterns of gene expression may still occur predominantly during the DNA/chromatin replication phase of the cell cycle.
In plants, deficiencies in methyltransferases impair epigenetic regulation, which is reflected by the release of transcriptional gene silencing (TGS) from various previously silent loci (Finnegan et al., 1996; Ronemus et al., 1996; Bartee et al., 2001; Lindroth et al., 2001). Since, with low methylating activity, passive demethylation would take place during DNA replication (Rougier et al., 1998), mitotic activity is an important factor for the release of silencing by this mechanism. However, a recent study of chromatin structure in quiescent Drosophila cells provided evidence that heterochromatic regions become accessible for transcription (Ahmad and Henikoff, 2001) and that histone replacement can take place independently of DNA replication (Ahmad and Henikoff, 2002), but the regulation of such potentially heritable chromatin changes at the level of the entire organism has not been investigated.
We have described a regulator of TGS, mom1 (Morpheus' Molecule 1), that releases TGS without alteration of DNA methylation (Amedeo et al., 2000). This suggests that MOM1 is involved in the recognition of other specific features of silent loci in the genome. MOM1 gene encodes a large nuclear protein with no overall similarity to other known proteins (Amedeo et al., 2000). To determine the mechanism of TGS control by MOM1, we examined whether MOM1 is required for the maintenance of TGS in non-dividing, differentiated cells. For this purpose, we established a chemically regulated MOM1 depletion system in Arabidopsis plants. The results presented here are consistent with the hypothesis that TGS maintenance mediated by MOM1 is a dynamic process relying on continuous supply/turnover of the MOM1 protein, rather than being linked to the faithful reproduction of epigenetic states during mitotic divisions.
Results and Discussion
The phenotype of TGS release caused by the mom1 mutation can be reproduced by the expression of MOM1 antisense RNA (Amedeo et al., 2000). Such transgenic modification of Arabidopsis led to the reactivation of transcriptionally silent transgenic loci and to the transcription of endogenous pericentromeric repeats, TSI (transcriptionally silent information), which are usually silent in wild-type Arabidopsis (Steimer et al., 2000). In order to inhibit MOM1 expression in differentiated cells after the extinction of their mitotic activity, we constructed a chemically inducible gene switch for MOM1 inhibition. We combined RNAi technology with the well-documented chemical activation of the PR-1 (pathogenesis-related 1) promoter of A. thaliana (Lebel et al., 1998). The PR-1 promoter is activated upon pathogen infection and can also be induced by a variety of exogenous chemical inducers, including benzo(1,2,3)thiadiazole-7-carbotioic acid S-methyl ester (BTH; Lawton et al., 1996). BTH systemically induces PR-1 promoter throughout the plant and therefore acts uniformly as the PR-1 inducer in all plant tissues except roots, where PR-1 is inactive (Lawton et al., 1996; Lebel et al., 1998).
The PR-1 promoter was linked to an inverted repeat (IR) construct derived from the 3′ end of MOM1 cDNA separated by the syn7 (synthetic 7) intron (Goodall and Filipowicz, 1989) (PR1-IRMOM; Figure 1A). The analogous RNAi construct was also coupled to the viral CaMV35S (Cauliflower Mosaic Virus) promoter (35S-IRMOM; Figure 1B) to compare induced with constitutive expression. Both constructs and empty vector (Figure 1C) as a control were stably introduced into Arabidopsis line 6b5 containing transcriptionally silent copies of a β-glucuronidase (GUS) transgene (Morel et al., 2000). GUS protein activity can be quantified fluorometrically and, as a cell-autonomous marker, can be localized histochemically throughout the entire plant. To consider the possible variation associated with individual transgenic lines expressing PR1-IRMOM or 35S-IRMOM, we examined several primary transgenic plants for alleviation of silencing. Transgenic plantlets grown in aseptic conditions for 24 days were transferred for a further 10 days to media with or without BTH (3 p.p.m.) and subsequently assayed for GUS activity. The activity was measured fluorometrically in extracts from individual plants, and the combined data are presented in Figure 2. Line 6b5 transformed with the control vector (no inverted repeat MOM; Figure 1C) maintained silencing of the GUS locus without or with BTH treatment (Figure 2). This indicates that BTH had no direct effect on TGS. On the other hand, TGS of the GUS locus could be clearly released in 35S-IRMOM transgenics without or with BTH treatment (Figure 2), which indicates the effectiveness of constitutively produced RNAi against MOM1. Most importantly, the release of silencing in populations of plants transgenic with PR1-IRMOM occurred only after treatment with BTH (Figure 2) to a similar extent as measured after constitutive expression of IRMOM (Figure 2).
Figure 1.
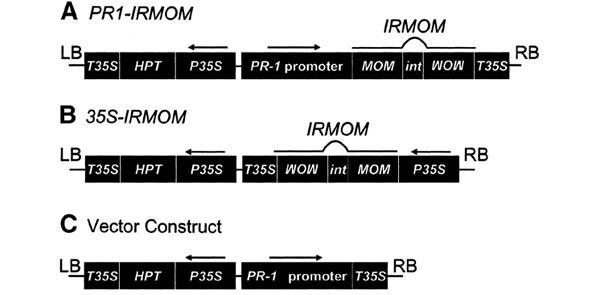
Functional maps of constructs. Inverted repeat MOM (IRMOM) was inserted behind (A) the PR-1 promoter (PR1-IRMOM) and (B) the CaMV 35S promoter (35S-IRMOM). (C) The control plasmid containing the PR-1 promoter but without IRMOM. Arrows above the promoters indicate direction of transcription; RB, right border; LB, left border; P35S, CaMV 35S promoter; T35S, 35S polyadenylation signal; HPT, hygromycin phosphotransferase gene; int, synthetic intron syn7 (Goodall and Filipowicz, 1989).
Figure 2.
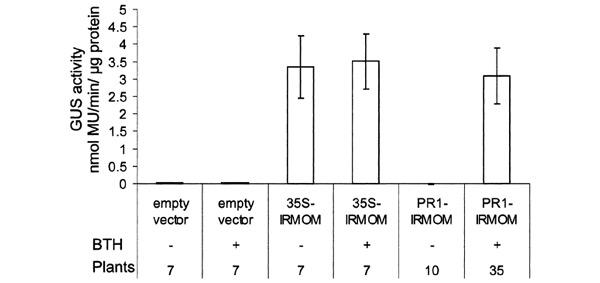
Levels of GUS activity in populations of primary transformants with and without BTH treatment. Transgenic plants (T1) transformed with the vector control, 35S-IRMOM or PR1-IRMOM were treated with 3 p.p.m. BTH (+) or mock treated (−). GUS activity was measured fluorometrically as described in the text and in Methods. The numbers of plants examined are indicated below the graph. Bars represent standard errors. Note: values and the error bars for the vector control treated with BTH or mock treated and PR1-IRMOM (without BTH) plants are too small to be depicted clearly.
In addition to the transgenic GUS locus of line 6b5, we also examined transcriptional reactivation of silent pericentromeric repeats termed TSI (Steimer et al., 2000). Expression of TSI was observed in 35S-IRMOM and PR1-IRMOM plants treated with BTH but not in plants transformed with vector or in PR1-IRMOM plants without BTH (Figure 3A). Accordingly, MOM1 transcript was depleted almost to undetectable levels in PR1-IRMOM plants treated with BTH and 35S-IRMOM plants but not in PR1-IRMOM plants without BTH or BTH-treated plants transformed with vector control (Figure 3B). Therefore, the induced expression of MOM1 RNA forming doublestranded structure very efficiently interferes with transcriptional silencing at transgenic and endogenous loci and is suitable for the histochemical determination of TGS release.
Figure 3.
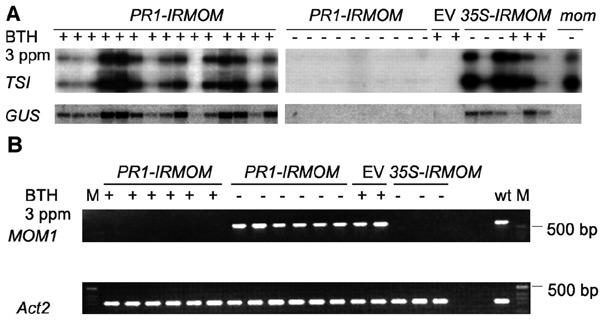
(A) Northern blot analysis of total RNA isolated from different individual primary transformants. PR1-IRMOM, empty vector (EV) and 35S-IRMOM transgenics were treated with (+) or without (−) BTH as described in Methods. mom mutant (without GUS locus; Amedeo et al., 2000) RNA was used as a control (last lane). The same blot was probed with TSI, GUS and cDNA of constitutively expressed RAN gene (Ras related nuclear protein; Haizel et al., 1997) as a loading control. (B) RT–PCR detection of MOM1 transcript. Total RNA isolated from individual primary transformants was reverse transcribed and PCR amplified with MOM1 specific primers and Act2 (Actin2) primers as described in Methods. M, marker; wt, wild type.
Histochemical detection of GUS expression after IRMOM expression in line 6b5 was performed in two complementary experimental set-ups. The first was analogous to that used for the quantification of GUS activity and the determination of mRNA levels of reactivated loci: 24-day-old plants were transferred for a further 10 days to medium supplemented with BTH before histochemical analysis of GUS expression. In the second set-up, 7-week-old plants with fully developed rosettes were sprayed with 200 p.p.m. BTH, and mature leaves were analysed 10 days later.
In both types of experiment, histochemical detection of GUS expression visualized TGS release only in 35S-IRMOM transgenic plants or in plants transformed with PR1-IRMOM after induction with BTH (Figure 4). Reactivation of the silent GUS locus was never observed in PR1-IRMOM plants without BTH induction, and BTH treatment itself did not interfere with TGS in control plants transformed with an empty vector (Figure 4A). No reactivation of the GUS locus was detected in the roots of PR1-IRMOM plants (Figure 4A; data not shown), which is in accordance with the absence of PR-1 expression in roots (Lebel et al., 1998).
Figure 4.
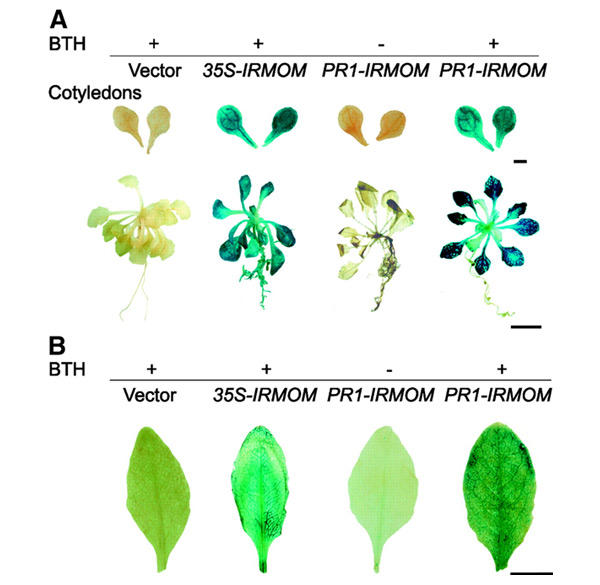
Histochemically determined GUS activity resulting from the release of silencing. (A) Twenty-four-day-old transgenic plants (T1) containing vector construct only, 35S-IRMOM or PR1-IRMOM were grown aseptically and treated with 3 p.p.m. BTH (+) or mock treated (−) as described in Methods. Upper: cotyledons; scale bar, 1 mm. Lower: whole plants, scale bar, 1 cm. (B) Forty-nine-day-old primary transformants (T1) for vector construct only, 35S-IRMOM or PR1-IRMOM were grown in soil and induced by spraying with 200 p.p.m. BTH (+). PR1-IRMOM were also mock treated (−). Leaves were stained for GUS activity as described in Methods. Scale bar, 1 cm. Note: mock-treated vector control and 35S-IRMOM plants are not shown, as they were exactly the same as BTH-treated plants.
Since reactivation of the GUS locus was clearly visible in cotyledons of 24-day-old plants (Figure 4A), which contain no mitotically or endomitotically active cells (De Veylder et al., 2002), and BTH does not activate genes involved in cell cycle progression (Maleck et al., 2000), this provided the first indication that the release of TGS by interference with MOM1 function may be independent of mitotic DNA replication. To re-examine this result, we determined patterns of TGS release also in mature Arabidopsis leaves, where zones of mitotic activity in the course of leaf development were studied in great detail. The major mitotic activity takes place in very young leaves before unfolding. In leaves of ∼3–3.5 mm, mitotic activity is largely confined to the base of the blade and there is no island of dividing cells in the upper part of the leaf. When leaves reach ∼8–8.5 mm, residual mitotic activity was only observed at the very base of the leaf and in the petiole (Pyke et al., 1991; Donnelly et al., 1999; De Veylder et al., 2001). However, the arrest of cell division activity does not mean the arrest of DNA replication, and endoreduplication is well described for many cells in Arabidopsis leaves (Joubes and Chevalier, 2000). The pattern of endoreduplication during early leaf development follows this basiplastic pattern of mitotic divisions (Jacqmard et al., 1999; Castellano et al., 2001) and occurs in a patchy fashion. In the later developmental stages there is no mitotic or endomitotic activity and the mature size of leaves is reached exclusively by the expansion of existing cells (Pyke et al., 1991; Donnelly et al., 1999; De Veylder et al., 2001). Obviously, if the release of silencing due to induced depletion of MOM1 is coupled to mitotic or endomitotic DNA replication, the pattern of TGS release should coincide with DNA replication sites. This is evidently not the case, since chemically regulated inhibition of MOM1 expression did not follow a basiplastic pattern of actively dividing cells in early leaf development and was effective in leaves that had clearly passed the stage of mitotic activity (Figure 4A).
To strengthen this observation, we applied BTH (sprayed with 200 p.p.m.) to 7-week-old plants with fully developed rosettes and determined GUS activity 10 days later. The results confirmed that it was also possible to alleviate TGS by downregulation of MOM1 expression in mature Arabidopsis plants at the beginning of bolting, i.e. after termination of rosette development (Figure 4B). Since the reactivated GUS transgene was regulated by CaMV 35S promoter (Morel et al., 2000), GUS activity was localized mainly in the vicinity of vascular bundles, reflecting patterns of 35S promoter expression in mature plants (Wilkinson et al., 1997). In conclusion, the observed patterns of TGS release in cotyledons and in rosette leaves of Arabidopsis plants after induced MOM1 depletion are consistent with the notion that MOM1 protein is required for TGS maintenance in cells that have stopped DNA replication.
Such properties of MOM1 resemble yeast SIR2 and SIR3 proteins as dynamic components of silent chromatin at mating-type loci (Miller and Nasmyth, 1984; Cheng and Gartenberg, 2000; Bedalov et al., 2001). Depletion of the SIR3 gene product by the utilization of its temperaturesensitive form or its overexpression can alter the silencing status of the mat loci in the interphase or on excised, non-replicating DNA molecules (Miller and Nasmyth, 1984; Cheng and Gartenberg, 2000). Chemical inhibition of SIR2 activity resulted in similar alleviation of silencing, suggesting that the SIR complex is constantly required for the maintenance of silencing in stationary cells (Bedalov et al., 2001).
In Drosophila, the only multicellular eukaryote studied in this respect, cell cycle progression is also not necessary to alter the silent epigenetic state, however by forced, local activation of heterochromatic region using GAL4 activator (Ahmad and Henikoff, 2001). Such competition between overexpressed transcription factor and repressive chromatin at the white locus revealed that Drosophila heterochromatin is in a dynamic state and that transcriptional activators can take advantage of transient accessibility of genes residing within heterochromatin. However, the endogenous molecular components of this dynamic heterochromatin in differentiated cells are not known. Our results suggest that MOM1 in plants serves this function and its availability can rapidly influence epigenetic states in non-dividing cells. In general, our results illustrate that the epigenetic make-up of differentiated cells within plant tissues, and possibly in tissues of other multicellular organisms, can be altered rapidly by the presence of particular TGS components. Such regulation can thus be explored both in nature and through biotechnology.
Methods
Plant material.
Seeds of Arabidopsis line 6b5 (Morel et al., 2000) and T1 transgenics were sterlized and grown in sterile culture or in soil under conditions described previously (Steimer et al., 2000).
Chemical activation of the PR-1 promoter.
BTH in a formulation containing 50% active ingredient in wet able powder (provided by U. Neuenschwander, Syngenta) was used for induction. After 24 days of growth on selective medium, primary transformants were transferred to liquid germination medium (Murashige and Skoog, 1962) containing 3 p.p.m. BTH in 24-well microtiter plates. After 3 days, the old medium was replaced with fresh medium containing 3 p.p.m. BTH, and histochemical staining was performed 7 days later.
In the case of soil-grown plants, 49-day-old plants were sprayed with 200 p.p.m. BTH dissolved in water, whereas the control plants were mock treated. Four days later, the spray was repeated and GUS activity was determined after a further 6 days.
Fluorometric GUS activity assay.
GUS activity was determined in extracts of total cellular proteins using 4-methyl-umbelliferyl β-D-glucuronide (MUG, Sigma) as described previously (Jefferson et al., 1987). The fluorescence of 4-methylumbelliferone (MU) was determined using a Titertek Fluoroskan II ELISA plate reader (Flow Laboratories). Protein concentrations in plant extracts were determined in a Bradford assay (Bio-Rad), according to the manufacturer's instructions. GUS enzyme activity is expressed in nmol MU/min/μg protein.
Northern blot analysis.
Total RNA from primary transformants, induced 24 days after germination, was isolated using TRIZOL (Gibco BRL, Grand Island, NY, USA) according to the supplier's instructions. After standard gel separation and blotting, filters were hybridized as described previously (Church and Gilbert, 1984).
RT–PCR.
Five micrograms of total RNA was treated with RQ1 DNase (Promega) according to the manufacturer's instructions. The RNA was reverse transcribed as described previously (Steimer et al., 2000), and cDNA was amplified for 30 PCR cycles (94°C for 30 s, 55°C for 30 s and 72°C for 30 s) with primers specific for MOM1 (Amedeo et al., 2000) derived from regions 5902–5927 (CD29F) and 6530–6505 (Cla3R). The Act2 (Actin2) primers (see Supplementary data available at EMBO reports Online) were directed towards Act2 (An et al., 1996).
Histochemical localization of GUS activity.
Assays for GUS expression were performed on primary transformants using 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-gluc, Fluka) with the addition of potassium ferricyanide and potassium ferrocyanide, both at 5 mM, in staining buffer as described previously (Mascarenhas and Hamilton, 1992).
Supplementary Material
Supplementary data
Acknowledgments
We thank H. Vaucheret for the Arabidopsis line 6b5 and U. Neuenschwander for providing BTH. We also thank J.P. Jost, B. Hohn and O. Mittelsten Scheid for their helpful comments on the manuscript.
References
- Ahmad K. and Henikoff S. (2001) Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell, 104, 839–847. [DOI] [PubMed] [Google Scholar]
- Ahmad K. and Henikoff S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell, 9, 1191–1200 [DOI] [PubMed] [Google Scholar]
- Amedeo P., Habu Y., Afsar K., Mittelsten Scheid O. and Paszkowski J. (2000) Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature, 405, 203–206. [DOI] [PubMed] [Google Scholar]
- An Y.Q., McDowell J.M., Huang S., McKinney E.C., Chambliss S. and Meagher R.B. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J., 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Araujo F.D., Knox J.D., Szyf M., Price G.B. and Zannis-Hadjopoulos M. (1998) Concurrent replication and methylation at mammalian origins of replication. Mol. Cell. Biol., 18, 3475–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L., Malagnac F. and Bender J. (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev., 15, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedalov A., Gatbonton T., Irvine W.P., Gottschling D.E. and Simon J.A. (2001) Identification of a small molecule inhibitor of Sir2p. Proc. Natl Acad. Sci. USA, 98, 15113–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano M.M., del Pozo J.C., Ramirez-Parra E., Brown S. and Gutierrez C. (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell, 13, 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.H. and Gartenberg M.R. (2000) Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev., 14, 452–463. [PMC free article] [PubMed] [Google Scholar]
- Church G.M. and Gilbert W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L. et al. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell, 13, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L. et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa–DPa transcription factor. EMBO J., 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E. and Dengler N.G. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol., 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Peacock W.J. and Dennis E.S. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl Acad. Sci. USA, 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G.J. and Filipowicz W. (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell, 58, 473–483. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. and Elgin S.C. (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev., 12, 178–187. [DOI] [PubMed] [Google Scholar]
- Haizel T., Merkle T., Pay A., Fejes E. and Nagy F. (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J., 11, 93–103. [DOI] [PubMed] [Google Scholar]
- Jacqmard A., De Veylder L., Segers G., de Almeida Engler J., Bernier G., Van Montagu M. and Inze D. (1999) Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta, 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A. and Bevan M.W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes J. and Chevalier C. (2000) Endoreduplication in higher plants. Plant Mol. Biol., 43, 735–745. [DOI] [PubMed] [Google Scholar]
- Kass S.U., Landsberger N. and Wolffe A.P. (1997) DNA methylation directs a time-dependent repression of transcription initiation. Curr. Biol., 7, 157–165. [DOI] [PubMed] [Google Scholar]
- Lawton K.A., Friedrich L., Hunt M., Weymann K., Delaney T., Kessmann H., Staub T. and Ryals J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J., 10, 71–82. [DOI] [PubMed] [Google Scholar]
- Lebel E., Heifetz P., Thorne L., Uknes S., Ryals J. and Ward E. (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J., 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S. and Jacobsen S.E. (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science, 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L. and Dietrich R.A. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet., 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J.P. and Hamilton D.A. (1992) Artifacts in the localization of GUS activity in anthers of petunia transformed with CaMV 35S–GUS construct. Plant J., 2, 405–408. [Google Scholar]
- Miller A.M. and Nasmyth K.A. (1984) Role of DNA replication in the repression of silent mating type loci in yeast. Nature, 312, 247–251. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain P., Beclin C. and Vaucheret H. (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol., 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog F. (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant, 15, 473–497. [Google Scholar]
- Pyke K.A., Marrison J.L. and Leech R.M. (1991) Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L) Heynh. J. Exp. Bot., 42, 1407–1416. [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J. and Dellaporta S.L. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science, 273, 654–657. [DOI] [PubMed] [Google Scholar]
- Rougier N., Bourc'his D., Gomes D.M., Niveleau A., Plachot M., Paldi A. and Viegas-Pequignot E. (1998) Chromosome methylation patterns during mammalian preimplantation development. Genes Dev., 12, 2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer A., Amedeo P., Afsar K., Fransz P., Mittelsten Scheid O. and Paszkowski J. (2000) Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell, 12, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J.E., Twell D. and Lindsey K. (1997) Activities of CaMV 35S and nos promoter in pollen: implications for field release of transgenic plants. J. Exp. Bot., 48, 265–275. [Google Scholar]
- Xu L., Glass C.K. and Rosenfeld M.G. (1999) Coactivator and corepressor complex in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


