Abstract
Calcium (Ca2+) signals regulate a diverse set of cellular responses, from proliferation to muscular contraction and neuro-endocrine secretion. The ubiquitous Ca2+ sensor, calmodulin (CaM), translates changes in local intracellular Ca2+ concentrations into changes in enzyme activities. Among its targets, the Ca2+/CaM-dependent protein kinases I and IV (CaMKs) are capable of transducing intraneuronal signals, and these kinases are implicated in neuronal gene regulation that mediates synaptic plasticity in mammals. Recently, the cyclic AMP response element binding protein (CREB) has been proposed as a target for a CaMK cascade involving not only CaMKI or CaMKIV, but also an upstream kinase kinase that is also CaM regulated (CaMKK). Here, we report that all components of this pathway are coexpressed in head neurons of Caenorhabditis elegans. Utilizing a transgenic approach to visualize CREB-dependent transcription in vivo, we show that this CaMK cascade regulates CRE-mediated transcription in a subset of head neurons in living nematodes.
Introduction
It is clear from overexpression studies that calcium/calmodulin (Ca2+/CaM)-dependent protein kinases (CaMKs) can influence the phosphorylation state and transcriptional transactivating activity of the cyclic AMP response element binding protein (CREB; Sun et al., 1994; Bito et al., 1996), but thus far the only evidence that CREB might be a target of a CaMK in vivo comes from the demonstration that CaMKIV-null mice exhibit reduced CREB phosphorylation in a subset of cells that includes neurons of the central nervous system (Ho et al., 2000; Ribar et al., 2000). Furthermore, although the components of a CaMK cascade (a CaMK kinase, CaMKK, and a CaMKI or CaMKIV) have been identified and characterized in several organisms, as yet there is no evidence that these constituents act as a cascade in vivo or that such a cascade has physiologic consequences. Recent studies identified sole Caenorhabditis elegans homologs of each of the putative cascade components, CaMKK (ckk-1; Tokumitsu et al., 1999) and CaMKI (cmk-1; Eto et al., 1999), and established that, in vitro, CKK-1 could activate CMK-1 through phosphorylation of Thr179. In addition, it was demonstrated that C. elegans kinases heterologously expressed in COS-7 cells promoted phosphorylation of mammalian CREB and CRE-dependent transcription (Eto et al., 1999). Those results suggested that CREB could be a target of a C. elegans CaMK cascade analogous to the presumptive mammalian CaMKK/CaMKIV cascade. Here, we identify the C. elegans homolog of CREB (crh-1) and demonstrate that a CaMKK/CaMK/CREB cascade functions in neurons in living nematodes.
Results and Discussion
To isolate a CREB homolog in C. elegans, we screened a cDNA library using as a probe a C. elegans cDNA project clone (yk217f19) that showed high homology to the C-terminal coding region of mouse CREB (encoding residues 296–337). A cDNA containing an open reading frame (ORF) predicted to encode a protein of 306 amino acids was isolated; the sequence shared ∼50% primary sequence identity with vertebrate CREBs (Gonzalez et al., 1989; Meyer and Habener, 1992; Sakaguchi et al., 1999). Importantly, both a kinase inducible domain (KID, residues 5–50) containing a putative activation phosphorylation site (Arg-Arg-Proser29, homologous to Ser133 in mammalian CREB), and a basic and leucine zipper domain (bZIP, residues 243–304, required for dimerization and DNA binding) are particularly well conserved (80 and 95% identity, respectively; Figure 1A). Several splice variants identified by 5′-RACE (rapid amplification of cDNA ends) incorporate small changes into the N-terminus without altering the integrity of the KID and bZIP domains and/or overall homology. However, none of these variants is identical to the predicted ORFs in this region, Y41C4A.4a and Y41C4A.4b. A survey of the C. elegans genome found at least 19 bZIP-containing genes (Ruvkun and Hobert, 1998), but crh-1 is unique in encoding both bZIP and KID motifs, indicating that it is the only CREB family gene in C. elegans.
Figure 1.
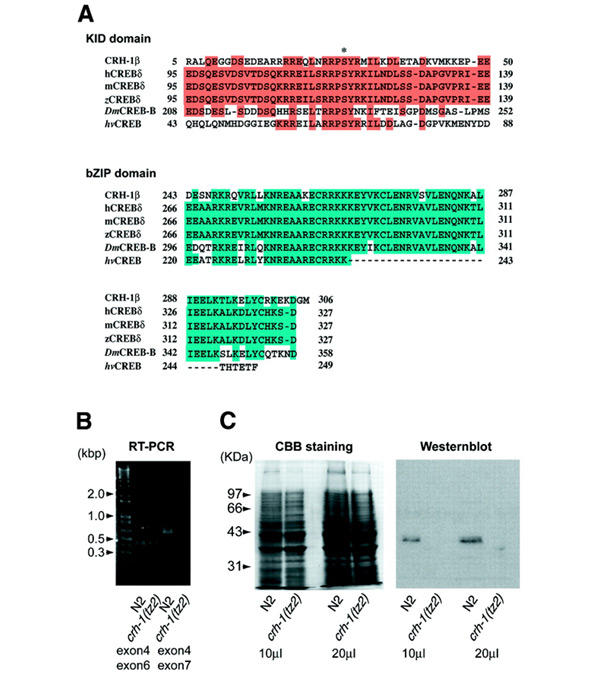
CREB homolog of C. elegans (crh-1). (A) An amino acid sequence alignment of KID and bZIP domains of the CREB family from C. elegans (CRH-1β), human (hCREB), mouse (mCREB), zebra finch (zCREB), Drosophila (DmCREB2-a) and hydra (hvCREB). Shading indicates identical amino acids. The asterisk indicates the putative phosphorylated Ser residue of each CREB family member. (B) Analysis of RT–PCR in N2 and crh-1(tz2) worms. cDNAs were amplified with primers from exons 4 and 6 (left) and 4 and 7 (right), respectively. Exon 7 is deleted in crh-1(tz2) strain. (C) Analysis of CRH-1 phosphorylation in N2 and crh-1(tz2) worms. Worm extracts (10 and 20 μl) were prepared with SDS–PAGE sample buffer and subjected to western blot analysis using anti-phospho-CREB antibody. CBB, Coomassie Brilliant Blue.
Through screening a library of mutagenized nematodes, we identified a strain, YT3 crh-1(tz2), carrying a deletion of 979 nucleotides (54730–55708 in Y41C4A; from the last exon 7 to the 3′ UTR of crh-1) that eliminates 38 amino acids at the C-terminal of the bZIP region (Glu268-Met306). RT–PCR showed that both N2 and crh-1(tz2) expressed mRNA until exon 6 of crh-1, but crh-1(tz2) does not express full-length mRNA for crh-1 (Figure 1B). Western blot analysis using anti-phospho-CREB antibody revealed that the N2 strain contains the expected immunoreactive 40 kDa band, whereas the 40 kDa band was not detected in crh-1(tz2) (Figure 1C), indicating that crh-1(tz2) could not produce functional CRH-1 proteins.
To confirm that the crh-1 gene product can be phosphorylated by the C. elegans CaMK, recombinant His-tagged CRH-1β was purified from bacteria and used as a substrate for in vitro kinase assays. As shown in Figure 2A, CRH-1β is weakly phosphorylated by CMK-1 alone, but the phosphate incorporation is markedly enhanced by the activation of CMK-1 with CKK-1, as assessed either by 32P-incorporation or by immunoreactivity with anti-phosphoser133 of mammalian CREB. This is consistent with previous observations that CKK-1 activates CMK-1 via phosphorylation of Thr179 and thereby decreases CMK-1's Km value for peptide substrates (Eto et al., 1999; Hook et al., 1999). Mutation of CRH-1β Ser29 to Ala completely abolished its phosphorylation by activated CMK-1, indicating that this site is the only CMK-1 phosphorylated residue.
Figure 2.
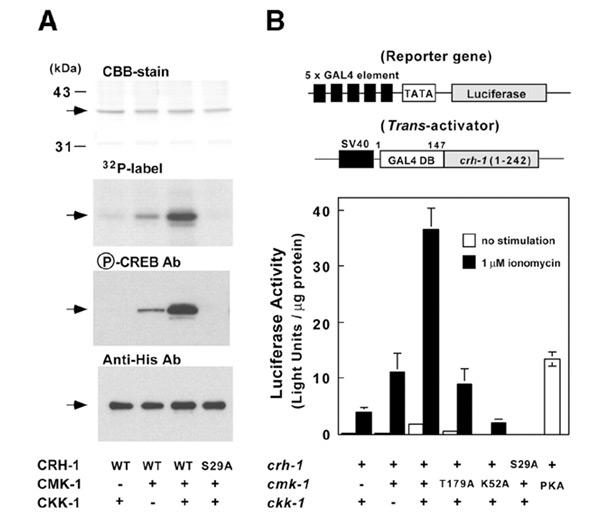
Phosphorylation and transcriptional regulation of CRH-1β by C. elegans CaMK cascade. (A) Either wild-type (WT) or Ser29Ala (S29A) mutant of CRH-1β (0.2 μg) was incubated with CMK-1 (22 ng) in either the absence or the presence of CKK-1 (0.5 ng) in a solution (25 μl) containing 30 mM HEPES (pH 7.5), 5 mM MgAc2, 1 mM DTT, 2 mM CaCl2, 3 μM CaM and either 100 μM [γ-32P]ATP or 100 μM cold ATP at 30°C for 10 min as described in Methods. After terminating the reaction by the addition of 5 μl of SDS–PAGE buffer, the sample (30 μl for autoradiography and 5 μl for western blot) was subjected to SDS 10% PAGE followed by either autoradiography or western blotting using anti-phospho-CREB antibody or anti-His-tag antibody. Arrows indicate CRH-1β. (B) COS-7 cells (6-well dishes) were transfected with 0.5 μg of a reporter gene (pFR-5 × GAL4-binding element-Luciferase) and expression plasmid (0.5 μg) carrying GAL-4 DNA-binding domain fused with residues 1–242 of either crh-1β wild-type crh-1β or crh-1β (Ser29Ala) and a combination of expression plasmids (pME18s) carrying either cmk-1 wild type or mutants (0.5 μg) and/or ckk-1 (0.5 μg) as indicated. A cDNA of the catalytic subunit of mammalian PKA (0.5 μg) was also used as a positive control. After incubation for 24 h, the cells were deprived of serum for 6 h and then stimulated with (filled bars) or without (open bars) 1 μM ionomycin for 12 h. Then the luciferase activity of each cell extract was measured as described in Methods. Results represent mean ± SEM of three independent transfections.
That phosphorylation of CRH-1β by the CaMK cascade is capable of activating transcription was shown using the GAL4-luciferase reporter assay system. The cDNA sequence encoding the transactivation domain of CRH-1β (1–242, lacking the bZIP domain) was fused to the DNA-binding domain of GAL4 and transfected into COS-7 cells along with a 5 × Gal luciferase reporter gene and various combinations of CMK-1 and CKK-1 expression plasmids (Figure 2B). Ionomycin-induced CRH-1β-dependent transcription was significantly enhanced by coexpression of wild-type CMK-1, in comparison with a kinase-inactive CMK-1 mutant, cmk-1 (K52A). Co-transfection of ckk-1 with wild-type cmk-1 further stimulated transcriptional activity 3- to 4-fold, an effect that was not apparent when the activation loop mutant of cmk-1, Thr179Ala, was substituted for wild-type cmk-1 in the assay. Furthermore, the kinases' effects were abolished by using the Ser29Ala mutant of CRH-1β, indicating that phosphorylation of this Ser is required for CRH-1β-dependent transcription. Taken together, these results indicate that Ca2+ mobilization activates the C. elegans CaMK cascade, which leads to phosphorylation and activation of CRH-1 in transfected cells.
We evaluated the expression patterns of the ckk-1, cmk-1 and crh-1 genes to establish that the presumptive cascade members are appropriately located to function as a signaling pathway in C. elegans. For each of the kinases, several kilobases of genomic sequence upstream of each coding region was used to promote GFP expression in transgenic nematodes. Adult hermaphrodites carrying a ckk-1::GFP fusion gene express GFP primarily in head and tail neurons and vulval muscles (Figure 3, top). Nematodes carrying the analogous cmk-1 vector exhibit strong fluorescence in numerous neurons and weak expression in vulval muscle cells (Figure 3, middle). crh-1 mRNA expression was sufficiently robust that we visualized its distribution by whole-mount in situ hybridization. In adult hermaphrodites, crh-1 expression is limited to head neurons and gonads (Figure 3, bottom). These crh-1 expression results were unexpected, as CREB is ubiquitously distributed in mammalian cells. Thus, only in a subset of head neurons of C. elegans are the components of this putative CaMK cascade poised to function in a linear signaling pathway.
Figure 3.
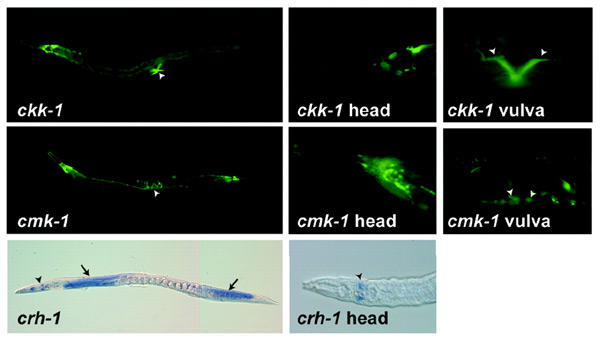
Expression patterns of ckk-1, cmk-1 and crh-1 in adult hermaphrodites. (Top) The expression pattern of ckk-1::GFP. Arrowheads indicate the expression in vulval muscles. (Middle) The expression pattern of cmk-1::GFP. Arrowheads indicate the expression in vulval muscles. (Bottom) The localization of crh-1 transcripts. Arrows and arrowheads indicate the expression in head neurons and germ cells in an ovary, respectively.
To examine whether a CKK-1/CMK-1/CRH-1 cascade operates in living nematodes, we monitored the regulation of CRE-driven transcription using nematodes carrying a pCRE::GFP monitoring vector (pYT39.93.1). Expression of GFP was undetectable or weakly detectable in wild-type (N2) nematodes injected with low concentrations (30 ng/μl) of this plasmid alone, although injection of higher concentrations (90 ng/μl) could induce weak fluorescence in a few pairs of neurons in head and tail (Figure 4A). Those animals in which wild-type cmk-1 was co-injected showed moderate fluorescence in comparison with those carrying only pCRE::GFP; 60% of the nematodes expressed GFP in a subset of the head neurons (1.2 ± 0.1 neurons/animal; N=110 transgenics; Figure 4B). On the other hand, >90% of transgenics expressing the truncated Ca2+/CaM-independent form of cmk-1 (1–295) exhibited intense GFP expression in head neurons (3.4 ± 0.4 neurons/animal; N=113) that was diminished by the Thr179Ala mutation in cmk-1 (1.1 ± 0.2 neurons/animal; N=101), suggesting that phosphorylation of this residue is required for CRE-dependent transcriptional upregulation in intact nematodes. Support for the stimulatory nature of this modification was provided through the use of a similarly truncated cmk-1 in which a phosphomimetic Asp residue replaced the activation loop Thr; this kinase enhanced GFP expression beyond that stimulated by cmk-1 (1–295). The role of activating CMK-1 was formally attributed to the endogenous CKK-1 by performing the in vivo reporter assay in a ckk-1-deficient strain, ckk-1(tm421) (Figure 4C). In contrast to the effects observed in the N2 strain, ckk-1-deficient nematodes carrying Ca2+/CaM-independent cmk-1 did not demonstrate stimulated transcription. As anticipated, fluorescence induced by the Thr179Asp form of cmk-1 was robust, even in the ckk-1-null genetic background. Taken together, these experiments show that cmk-1 affects CRE-transcriptional activation in a manner consistent with Ca2+/CaM binding and phosphorylation of Thr179 by CKK-1.
Figure 4.
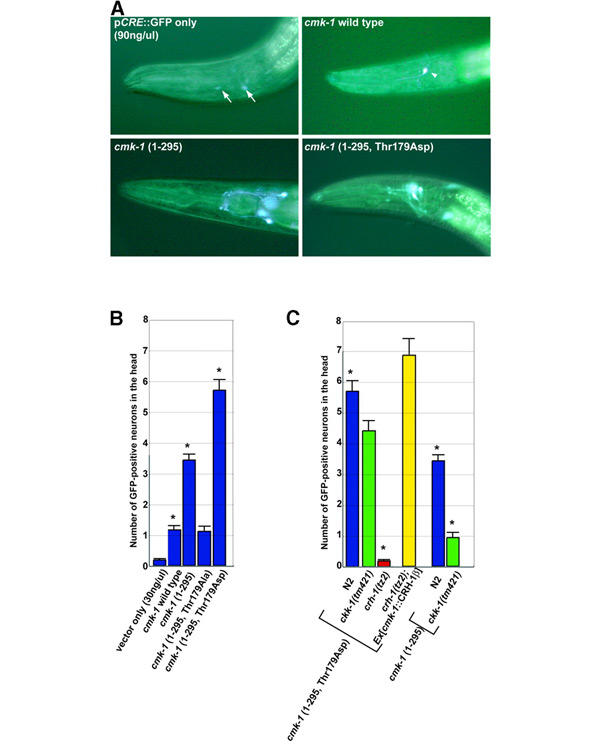
Monitoring CRE-dependent transcription in living nematodes. (A) GFP fluorescence of a transgenic worm carrying only pCRE::GFP (90 ng/μl), wild-type, Ca2+/CaM-independent (1–295) or constitutively active forms of cmk-1 (1–295, Thr179Asp). Arrows indicate the expression seen in neurons when a large amount of pCRE::GFP was injected. The arrowhead indicates the typical asymmetrical expression often seen in pharynx neurons when wild-type cmk-1 was injected. (B) Comparison of the number of fluorescent neurons in transgenic animals carrying only pCRE::GFP (30 ng/μl), wild-type, Ca2+/CaM-independent (1–295), unphosphorylated (1–295, Thr179Ala) or constitutively active (1–295, Thr179Asp) form of cmk-1 in wild-type animals. (C) Effect of crh-1 and ckk-1 genes for the enhanced expression induced by the constitutively active (1–295, Thr179Asp) or Ca2+-independent (1–295) forms of cmk-1. Error bars equal SEM. The p-value (asterisks, <0.0001) in (B) indicates a significant difference in CRE-GFP inducible activity between the wild-type and various forms of cmk-1. The p-value (asterisks, <0.0001) in (C) indicates a significant difference of fluorescence between N2 and either the crh-1 or the ckk-1 mutant.
As CREB is not the only transcription factor capable of binding CRE regulatory sequences, it was possible that crh-1 was not mediating the CaMK cascade's activation of CRE::GFP expression. However, when the cascade function was examined in the crh-1-deficient strain, crh-1(tz2), cmk-1 (1–295, Thr179Asp) did not stimulate expression of CRE::GFP, and crh-1β expression under the control of the cmk-1 promoter can completely rescue the CRE::GFP fluorescence in the crh-1-deficient strain (Figure 4C), indicating that crh-1 is indeed downstream of the CaMK cascade. Thus, we have provided direct genetic evidence that a CaMKK/CaMK/CREB cascade functions to activate CRE-dependent transcription in head neurons of C. elegans.
Using transgenic and mutant C. elegans strains as complementary tools, we have demonstrated transcriptional regulation in vivo by the kinase cascade predicted from in vitro enzyme and transfection assays. Yet, while CaMKK activates CaMK in neurons, the upstream kinase is expressed in only a subset of the other mammalian and C. elegans cell types that express a CaMK. This seems extravagant if phosphorylation by CaMKK is absolutely necessary for the activation of CaMK and suggests that there may be kinases other than CaMKK that are capable of activating CaMKI/CaMKIV (Soderling 1999). Alternatively, activation loop phosphorylation of the CaMK may not be required for its activity towards all substrates, as is indicated by in vitro peptide phosphorylation analyses (Hook et al., 1999). The ckk-1-deficient nematode should provide an ideal system in which to further discern the biochemical requirements for localized CaMK function.
Although our genetic analyses using pCRE::GFP demonstrate that the cascade activates the transcription of CRE-sequence-dependent genes through CRH-1, the physiological function of this cascade has not been assessed yet. Pathway components are coexpressed only in sensory neurons and interneurons in the head, suggesting that the CaMK cascade may regulate nematodes' responses to environmental stimuli. Actually, crh-1-dedicient worms looked normal and fertile; however, they grew slightly slower than the wild type (data not shown). In addition, our preliminary observation suggested that crh-1(tz2) worms have some behavioral defects. For example, they are slightly clumpy and tend to burrow into the agar plate even in food-rich conditions (data not shown), indicating that crh-1-deficient worms have some defects in sensing and/or integrating the stimuli from circumstances. However, we could not find any structural abnormalities in the sensory neurons visualized by the retrograde labeling with DiQ (data not shown). Several mutations in C. elegans genes whose mammalian homologs interact with CaMKK/CaMK/CREB signaling in biochemical and transfection experiments (CaMKII and ras/MAPK/MAPKK; Sun et al., 1994; Enslen et al., 1996) have developmental and behavioral phenotypes associated with dysfunction of these same neurons (CaMKII, unc-43; ras, let-60; MAPKKs, sek-1, nsy-1; Reiner et al., 1999; Sagasti et al., 2001; Hirotsu et al., 2000; Tanaka-Hino et al., 2002). The relative contributions of these pathways and the CaMK cascade in transducing Ca2+ signals in various contexts are unknown. Our genetic model system provides an opportunity to assess functional outcomes dependent on physiological interactions between the CaMK cascade and these other biochemical pathways and establishes a precedent for applying this direct in vivo approach to map endogenous signaling networks.
Methods
Transgenic strains generation and analysis.
Germline transformation was carried out as described previously (Mello et al., 1991; Mitani 1995). Host strains were either lin-15(n765ts), crh-1(tz2), ckk-1(tm421) or wild-type N2. The lin-15 clone pJMZ (10–50 ng/μl; Clark et al., 1994) was used as a co-injection marker for lin-15(n765ts). Transgenic lines were identified by rescue of the multivulva phenotype at 20°C. For the other mutant and wild-type strains, plasmid pRF4 (Rol) was used as a co-injection marker. All test DNA was injected at 30–100 ng/μl, except the pCRE::GFP plasmid, which was co-injected with cmk-1 plasmids at concentrations <30 ng/μl to minimize background fluorescence. A minimum of three independent lines were selected and analyzed for each DNA pool. Chromosomal integrated lines carrying pCRE::GFP; pCRE::GFP and cmk-1::CMK-1(T179D); cmk-1::GFP; ckk-1::GFP were established by UV crosslinking (Stratalinker1800, Stratagene) at a dose of 300 J/m2, then backcrossed twice before use.
Supplementary Material
Supplementary data
Acknowledgments
We thank Y. Kohaha for cDNA clones and the cDNA library, T. Ishihara for DiQ, A. Coulson for cosmid clones and A. Fire for GFP expression vectors. We also thank A. Arai, T. Kaneko, N. Takahashi, M. Ito, E. Mizuochi and K. Morinaka for technical support and K. Winkler for critical reading. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Center for Research Resources. This research was supported in part by Grant-in-Aid for Scientific Research (H.T.), Research for the Future Program (M.H.), the US Department of Defense (E.E.C.) and the NIH (A.R.M.).
References
- Bito H., Deisseroth K. and Tsien R.W. (1996) CREB phosphorylation and dephosphorylation: a Ca2+ and stimulus duration-dependent switch for hippocampal gene expression. Cell, 87, 1203–1214. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Lu X. and Horvitz H.R. (1994) The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics, 137, 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslen H., Tokumitsu H., Stork P.J., Davis R.J. and Soderling T.R. (1996) Regulation of mitogen-activated protein kinases by a calcium/calmodulin-dependent protein kinase cascade. Proc. Natl Acad. Sci. USA, 93, 10803–10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K., Takahashi N., Kimura Y., Masuho Y., Arai K., Muramatsu M.A. and Tokumitsu H. (1999) Ca2+/calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional activation. J. Biol. Chem., 274, 22556–22562. [DOI] [PubMed] [Google Scholar]
- Gonzalez G.A., Yamamoto K.K., Fischer W.H., Karr D., Menzel P., Biggs W. 3rd, Vale W.W. and Montminy M.R. (1989) A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature, 337, 749–752. [DOI] [PubMed] [Google Scholar]
- Hirotsu T., Saeki S., Yamamoto M. and Iino Y. (2000) The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature, 404, 289–293. [DOI] [PubMed] [Google Scholar]
- Ho N. et al. (2000) Impaired synaptic plasticity and cAMP response element-binding protein activation in Ca2+/calmodulin-dependent protein kinase type IV/Gr-deficient mice. J. Neurosci., 20, 6459–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook S.S., Kemp B.E. and Means A.R. (1999) Peptide specificity determinants at P-7 and P-6 enhance the catalytic efficiency of Ca2+/calmodulin-dependent protein kinase I in the absence of activation loop phosphorylation. J. Biol. Chem., 274, 20215–20222. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D. and Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T.E. and Habener J.F. (1992) Cyclic AMP response element binding protein CREB and modulator protein CREM are products of distinct genes. Nucleic Acids Res., 20, 6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S. (1995) Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhaabditis elegans. Dev. Growth Differ., 37, 551–557. [DOI] [PubMed] [Google Scholar]
- Reiner D.J., Newton E.M., Tian H. and Thomas J.H. (1999) Diverse behavioural defects caused by mutations in Caenorhabditis elegans unc-43 CaM kinase II. Nature, 402, 199–203. [DOI] [PubMed] [Google Scholar]
- Ribar T.J., Rodriguiz R.M., Khiroug L., Wetsel W.C., Augustine G.J. and Means A.R. (2000) Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J. Neurosci., 20, RC107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. and Hobert O. (1998) The taxonomy of developmental control in Caenorhabditis elegans. Science, 282, 2033–2041. [DOI] [PubMed] [Google Scholar]
- Sagasti A., Hisamoto N., Hyodo J., Tanaka-Hino M., Matsumoto K. and Bargmann C.I. (2001) The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell, 105, 221–232. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Wada K., Maekawa M., Watsuji T. and Hagiwara M, (1999) Song-induced phosphorylation of cAMP response element binding protein in the songbird brain. J. Neurosci., 19, 3973–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling T.R. (1999) The Ca2+-calmodulin-dependent protein kinase cascade. Trends Biochem. Sci., 24, 232–236. [DOI] [PubMed] [Google Scholar]
- Sun P., Enslen H., Myung P.S. and Maurer R.A. (1994) Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev., 8, 2527–2539. [DOI] [PubMed] [Google Scholar]
- Tanaka-Hino M., Sagasti A., Hisamoto N., Kawasaki M., Nakano S., Ninomiya-Tsuji J., Bargmann C.I. and Matsumoto K. (2002) SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO rep., 3, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumitsu H., Takahashi N., Eto K., Yano S., Soderling T.R. and Muramatsu M. (1999) Substrate recognition by Ca2+/Calmodulin-dependent protein kinase kinase. Role of the arg-pro-rich insert domain. J. Biol. Chem., 274, 15803–15810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


