Abstract
Several real-time PCR procedures for the detection and genotyping of oocysts of Cryptosporidium parvum were evaluated. A 40-cycle amplification of a 157-bp fragment from the C. parvum β-tubulin gene detected individual oocysts which were introduced into the reaction mixture by micromanipulation. SYBR Green I melting curve analysis was used to confirm the specificity of the method when DNA extracted from fecal samples spiked with oocysts was analyzed. Because C. parvum isolates infecting humans comprise two distinct genotypes, designated type 1 and type 2, real-time PCR methods for discriminating C. parvum genotypes were developed. The first method used the same β-tubulin amplification primers and two fluorescently labeled antisense oligonucleotide probes spanning a 49-bp polymorphic sequence diagnostic for C. parvum type 1 and type 2. The second genotyping method used SYBR Green I fluorescence and targeted a polymorphic coding region within the GP900/poly(T) gene. Both methods discriminated between type 1 and type 2 C. parvum on the basis of melting curve analysis. To our knowledge, this is the first report describing the application of melting curve analysis for genotyping of C. parvum oocysts.
Cryptosporidium parvum is a coccidian protozoan that causes self-limited diarrhea in immunocompetent individuals. In immuncompromised patients and malnourished children the disease is severe, prolonged, and life threatening. Although several immunological and molecular methods for detection of C. parvum oocysts in stool and environmental samples have been developed (1, 11, 17, 34), immunomagnetic capture methods (8) have found widespread application, particularly for water monitoring (9, 29). The detection limits achieved with these systems are typically less then 10 oocysts, although recoveries are affected by the complexity of the matrix from which the oocysts are extracted. User-friendly molecular methods for the detection of oocysts in complex mixtures or genotyping of purified oocysts are needed for clinical and epidemiological applications and for water monitoring. Since no specific chemotherapy is available for this organism (7, 13), early detection of C. parvum infections, particularly in immunosuppressed patients and children, may be critical to provide supportive treatment. Furthermore, the detection of asymptomatic individuals and animals excreting oocysts may be helpful for preventing secondary infections, studying transmission routes, and identifying reservoir hosts.
More than 10 years have passed since the first report describing the detection of C. parvum by PCR (20). PCR has also been integrated into various genotyping procedures, such as restriction fragment length polymorphism analysis (31, 35, 37), random amplification methods (23), and methods detecting conformational polymorphisms (12). These approaches have been instrumental in advancing our understanding of the taxonomy of the genus Cryptosporidium (24, 38) and for studying the transmission of Cryptosporidium species and genotypes between various host species (22, 39).
More recently, a TaqMan real-time PCR assay (14) for the detection of C. parvum based on the amplification of an 835-bp sequence from the small-subunit rRNA was developed (15). Although the internal probe used in this assay was complementary to a polymorphic region of this gene, the assay did not discriminate among Cryptosporidium genotypes originating from various host species. The anonymous C. parvum locus originally identified by Laxer et al. (20) was chosen for a TaqMan assay by Guilot and Fontaine (E. Guilot and M. Fontaine, Abstr. Water Quality Technol. Conf., 2001). The limit of detection for this assay was five oocysts spiked into water pellets. The ability of real-time PCR to provide quantitative information was used by Krüger et al. (18) to develop an assay to quantitate the amount of C. parvum DNA present in a sample. Finally, a SYBR Green I-based real-time PCR assay (28) targeting the heat shock protein 70 locus of C. parvum was used to develop a commercial C. parvum detection assay (J. W. Czajka, G. D. DiGiovanni, M. E. Schaffer, A. M. Stolzfus, H. K. White, and M. W. LeChevallier, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. 2-1062, 2000). As with the TaqMan assays, these assays do not differentiate between C. parvum type 1 and type 2.
Here we report on the development of real-time PCR assays capable of detecting low numbers of oocysts and genotyping C. parvum. Polymorphic alleles were differentiated by SYBR Green I and fluorescent probe melting curve analysis (MCA) on the basis of single-nucleotide polymorphisms (SNPs).
MATERIALS AND METHODS
Parasites.
The isolates of C. parvum used in this study are listed in Table 1. Oocysts were purified from fecal samples as described previously (37).
TABLE 1.
Isolates of C. parvum tested
| Type | Isolate | Origin, location | Reference |
|---|---|---|---|
| 1 | TU502 | Human, United Statesa | Unpublished |
| UG502 | Human, Uganda | 10 | |
| UG405 | Human, Uganda | 10 | |
| UG1544 | Human, Uganda | Unpublished | |
| UG1259 | Human, Uganda | Unpublished | |
| UG1610 | Human, Uganda | Unpublished | |
| CDC728 | Human, United States | Unpublished | |
| 2 | GCH1 | Human, United States | 33 |
| OHIO | Bovine, United States | Unpublished | |
| TAMU | Equine, United States | 25 | |
| JRLHIV | Human, United States | Unpublished | |
| MD | Cervine, Scotland | 19 | |
| 7Cb | MDxUG405 recombinant | 10 | |
| UC Davis | Raccoon, United States | Unpublished |
Derived from UG502 by accidental human infection.
Recombinant laboratory isolate obtained by crossing type 2 isolates MD and UG405.
Oocyst micromanipulations and DNA extraction.
Individual oocysts of isolate GCH1 (32) were collected with a mouth-controlled pipette (16). Oocysts were suspended in water at a concentration of 104/ml. A 20-μl portion of this suspension was deposited onto a microscope slide, and the oocysts were viewed on an inverted microscope at ×200 magnification. Individual oocysts were aspirated into a manually pulled glass micropipette with a tip with an inner diameter of approximately 15 μm. Individual oocysts were deposited into microcentrifuge tubes containing 10 μl of PCR-grade water. Oocyst samples were then subjected to three cycles of freezing-thawing and were spun at 16,000 × g for 30 s. The supernatant was introduced directly into 20-μl capillaries together with the PCR premixture for PCR amplification. Alternatively, following freezing-thawing, DNA was extracted from the oocyst lysate with a High Pure PCR template preparation kit (Roche Diagnostics, Mannheim, Germany), as recommended by the manufacturer, except that 50 μl (instead of 200 μl) of elution buffer was used. For negative controls, volumes of 0.5 to 1 μl of water containing no visible oocysts were collected from the oocyst suspension and deposited into microcentrifuge tubes containing 10 μl of PCR-grade water and processed as described above for the oocysts.
To test the detection limit of the PCR method with oocysts isolated from feces, portions of 0.5-g fecal samples devoid of visible oocysts by microscopic examination of acid-fast-stained fecal smears (21) were spiked with 3,000 oocysts of isolate GCH1 obtained by serial dilution from a stock suspension of 3 × 106 oocysts/ml. Spiked fecal samples were homogenized in 5 ml of 0.9% saline and filtered through gauze to remove large particles. The fecal filtrates were then centrifuged at 800 × g for 5 min. The pellets were resuspended in 600 μl of 0.9% saline solution and divided into three equal parts of 200 μl each. DNA was isolated from individual aliquots by one of the following three methods: (i) by phenol-chloroform extraction followed by ethanol precipitation, (ii) with the High Pure PCR template preparation kit (Roche Diagnostics), and (iii) and with the Magna Pure LC total nucleic acid isolation kit (Roche Diagnostics). In all three methods DNA was recovered in 200 μl of elution buffer. Portions of 1 μl of DNA solution, theoretically equivalent to five oocysts, were used in each PCR mixture.
Real-time PCR with SYBR Green I.
PCR amplifications were performed in 20-μl capillary tubes with a LightCycler instrument (Roche Diagnostics). Reaction mixtures contained 1× LC-Fast Start DNA master mixture for SYBR Green I (Roche Diagnostics), 4 mM MgCl2, 10 pmol each of the forward and reverse primers, and 1 to 10 μl of DNA template. The tubes were capped, centrifuged at 700 × g for 5 s, and placed into the LightCycler carousel. Thermal cycling was performed as follows. A denaturation step of 10 min at 95°C was followed by 40 cycles of 0 to 1 s at 95°C, 4 to 5 s at the respective annealing temperature, and 7 to 22 s (depending on the template) at 72°C. Following amplification, the PCR products were identified by MCA by raising the temperature from 45 to 95°C at a rate of 0.05°C/s. During the initial optimization phase, following MCA, PCR products were analyzed on agarose gels to ensure that products of the correct size were amplified. To this aim, products amplified on the LightCycler instrument were recovered from the capillaries, mixed with loading buffer, and loaded onto 1.5% agarose gels in 0.5× TBE (Tris-borate-EDTA) buffer. The gels were stained with GelStar (FMC, Vallensbaek Strand, Denmark).
The following PCR primers were used: for β-tubulin protocol 1, primer btub1 (ATGCTGTAATGGATGTAGTTAGACA; positions 552 to 576 [GenBank accession no. Y12615]) (unpublished) and primer btub2 (GTCTGCAAAATACGATCTGG; positions 708 to 689) (36); for the poly(T) protocol, primer cry44 (CTCTTAATCCAATCATTACAAC; positions 1331 to 1352 [GenBank accession no. U83169]) and primer cry39 (GAGTCTAATAATAAACCACTG; positions 1649 to 1629) (5).
Real-time PCR with internal probes.
Oligonucleotides directed at the β-tubulin gene of C. parvum (GenBank accession no. Y12165) were synthesized by TIB Molbiol, Berlin, Germany. A 157-bp fragment was amplified from the β-tubulin gene exon 2 with forward primer btub1 and reverse primer btub2. Two negative-stranded internal probes, btubLC (positions 639 to 616) and btubFL (positions 665 to 641), were included in the reaction mixture at concentrations of 0.5 pmol each. Probe btubLC was labeled at the 5′ end with Red640, and probe btubFL was labeled at the 3′ end with fluorescein. The 3′ end of probe btubLC was blocked with a phosphate group to prevent extension by the polymerase. Fluorescence at 640 nm was generated by fluorescence resonance energy transfer following the annealing of both probes to their adjacent complementary sequences and the juxtaposition of the fluorescein and Red640 fluorophores. The sequences of the probes were identical to the β-tubulin sequence found in C. parvum type 2 (4, 30, 36). Discrimination between the type 1 allele and the type 2 allele was based on the presence of three SNPs, two within btubFL and one within btubLC (T→C at position 616, G→A at position 646, and C→T at position 662 [type 2→type 1 changes]). These mismatches reduced the affinity of the probes for the type 1 allele, thus lowering the melting temperature. PCR cycling conditions were as described above for the SYBR Green I method, except that for MCA the temperature was increased at a rate of 0.5°C/s and the reaction was run in 1× LC-Fast Start DNA master mixture for hybridization probes (Roche Diagnostics). The dynamic range and reproducibility of the assay were tested with 10-fold dilutions of C. parvum DNA corresponding to 104 oocysts to 1 oocyst per reaction mixture. The samples containing low numbers of oocysts were tested in triplicate.
RESULTS
Sensitivity of SYBR Green I real-time PCR method.
In a preliminary comparison, two protocols amplifying different fragments of the β-tubulin gene of C. parvum were evaluated. It was determined that the method targeting a 157-bp fragment flanked by primers btub1 and btub2 detected single oocysts, whereas a minimum of 1,000 oocysts was required for the other method. The sensitivity of the method with btub1-btub2 was further investigated with small numbers of oocysts introduced directly into the PCR mixture with a glass micropipette or with DNA extracted from oocysts collected individually by the same method. Table 2 summarizes the results from 59 experiments with oocysts collected individually with a micropipette or with negative control samples devoid of oocysts. A total of 37 reaction mixtures were spiked with one, two, three, or five oocysts and 22 reaction mixtures were spiked with blank samples, as shown in the spiked reaction and negative control columns, respectively, in Table 2. A majority (12 of 20) of the reaction mixtures spiked directly with single oocysts and half (3 of 6) of those spiked with DNA extracted from single oocysts gave a visible amplification product by PCR with btub1-btub2 (Fig. 1). Reaction mixtures spiked with DNA extracted from three and five oocysts were positive in three of four trials and three of three trials, respectively. Reaction mixtures spiked with DNA extracted from two oocysts were positive in two of four attempts. To assess the accuracy of the microisolation procedure and specifically to control for the possibility that additional oocysts or DNA may have inadvertently been introduced into the reaction mixtures, negative control samples were collected with the same micropipette system. Portions of 0.5 to 1 μl of water without visible oocysts were aspirated from the same experimental oocyst suspension. In the first series of assays, 3 of 12 negative control reaction mixtures amplified a detectable amplicon, suggesting either that oocysts were inadvertently collected and transferred to the PCR mixture or that DNA in solution was detected by the assay. To distinguish between these two possibilities, the oocysts from the experimental sample were precipitated by centrifugation and the supernatant was removed and replaced with the same volume of fresh PCR-grade water. Samples of 0.5 to 1 μl containing no visible oocysts were again aspirated from this washed oocyst suspension and subjected to PCR analysis. As shown in Table 2, none of these control reactions produced detectable PCR products. We conclude that the real-time PCR method with primers btub1 and btub2 can detect individual oocysts in a 40-cycle amplification reaction. The negative control experiments demonstrate that microisolation is an accurate method for the isolation of individual oocysts and that DNA present in solution may lead to false-positive results.
TABLE 2.
Real-time PCR detection of oocysts isolated by micromanipulation
| Test | No. of reaction mixtures spiked with the indicated no. of oocysts:
|
||||||
|---|---|---|---|---|---|---|---|
| Spiked reaction mixtures
|
Negative controls
|
||||||
| No DNA extraction
|
DNA extraction (spin column)
|
Unwashed oocyst suspension (0 oocysts) | Washed oocyst suspension (0 oocysts) | ||||
| One oocyst | Three oocysts | Five oocysts | One oocyst | Two oocysts | |||
| Replicate tests | 20 | 4 | 3 | 6 | 4 | 12 | 10 |
| Positive tests | 12 | 3 | 3 | 3 | 2 | 3 | 0 |
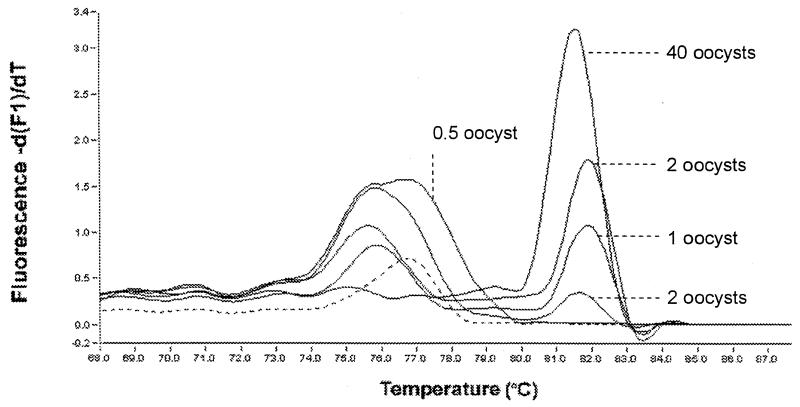
FIG. 1.
Detection of C. parvum oocysts recovered by micromanipulation. A diagnostic melting peak was detected with DNA extracted from 1, 2, and 40 oocysts with the High Pure PCR template preparation kit (Roche Diagnostics). The template with 0.5 oocyst was obtained by eluting DNA from 10 oocysts with 50 μl of elution buffer and introducing 2.5 μl of eluted DNA into a PCR mixture. In addition to the diagnostic melting peak, low-temperature melting peaks were observed, particularly when DNA from a few oocysts was amplified. These peaks may originate from primers-dimers, but their origins were not investigated. The dashed line shows the melting profile of the negative control reaction. Note that peak height is not always reflective of the amplicon concentration but is a measure of the slope of fluorescence versus temperature.
Specificity of real-time PCR with primers btub1 and btub2.
The specificity of the btub1-btub2 PCR method was investigated by adding DNA from the following protozoa into standard PCR mixtures: Entamoeba histolytica, Leishmania tropica, Toxoplasma gondii, and Giardia lamblia. DNA from none of these protozoa gave detectable amplification products with primers btub1 and btub2.
The specificity and the sensitivity of the btub1-btub2 amplification method were also tested with DNA extracted from C. parvum-negative fecal samples and fecal samples spiked with C. parvum oocysts. DNA was extracted from these samples by three methods: (i) by phenol-chloroform extraction followed by ethanol precipitation, (ii) with the High Pure DNA extraction kit (Roche Diagnostics), and (iii) with the Magna Pure DNA extraction kit (Roche Diagnostics). C. parvum DNA equivalent to that from five oocysts was detected in fecal samples from which DNA was extracted by all three methods, as demonstrated by the presence of a diagnostic melting peak at 81°C. DNA was also amplified from unspiked fecal samples, but MCA showed that the amplicons were distinct from those originating from C. parvum. We conclude from the results of these experiments that the assay with primers btub1 and btub2 detects low numbers of oocysts in feces. Amplification of non-C. parvum DNA either was not observed or was easily distinguished from the diagnostic amplicon by MCA.
Differentiation between C. parvum type 1 and type 2 by real-time PCR and with fluorescent probes.
A pair of probes 24 and 25 bp in length, respectively, were tested in a real-time PCR format with the same btub1 and btub2 amplification primers used for the SYBR Green I method. The purpose of this assay was to design a real-time PCR method capable of differentiating between C. parvum type 1 and type 2. The probes were designed such that their recognition sequence included multiple SNPs between the β-tubulin alleles found in type 1 and type 2. In this assay, fluorescence originated from fluorescence resonance energy transfer between a fluorescein and a Red640 fluorophore present on the 3′ end of the upstream btubFL probe and the 5′ end of the downstream btubLC probe, respectively. As shown in Fig. 2, MCA differentiated between C. parvum type 1 and type 2. As expected from the sequence identity of the probes with the type 2 β-tubulin allele, the melting temperature of type 2 isolates was higher than that of type 1 isolates. The presence of two SNPs within the btubFL recognition sequence decreased the affinity of the probe for the type 1 β-tubulin allele by an average of 4.9°C. In order to assess the reproducibility of this method for the detection of C. parvum type 1 and type 2, a total of 10 independent experiments were conducted over a period of 5 months. In each experiment the melting temperature of the probes was determined with the btubS and btub2 amplification primers and DNA from isolate GCH1 (type 2) and isolate TU502 (type 1). The mean melting temperatures were 65.27°C (standard deviation [SD] = 1.82; n = 10) for type 2 and 60.33°C (SD = 0.88; n = 11) for type 1. The difference in melting temperatures between type 1 and type 2 was statistically highly significant by the t test (P < 0.001). Several natural and laboratory type 2 isolates (isolates GCH1, TAMU, and OHIO) and type 1 isolates (isolates UG1544, UG1259, and UG1610) were tested by this method (Fig. 2). The melting temperature among type 1 isolates was more homogeneous than that among type 2 isolates, but the origin of this heterogeneity was not further investigated. The results of gel electrophoretic analysis of the amplicons from seven C. parvum isolates obtained with primers btub1 and btub2 are shown in Fig. 3.
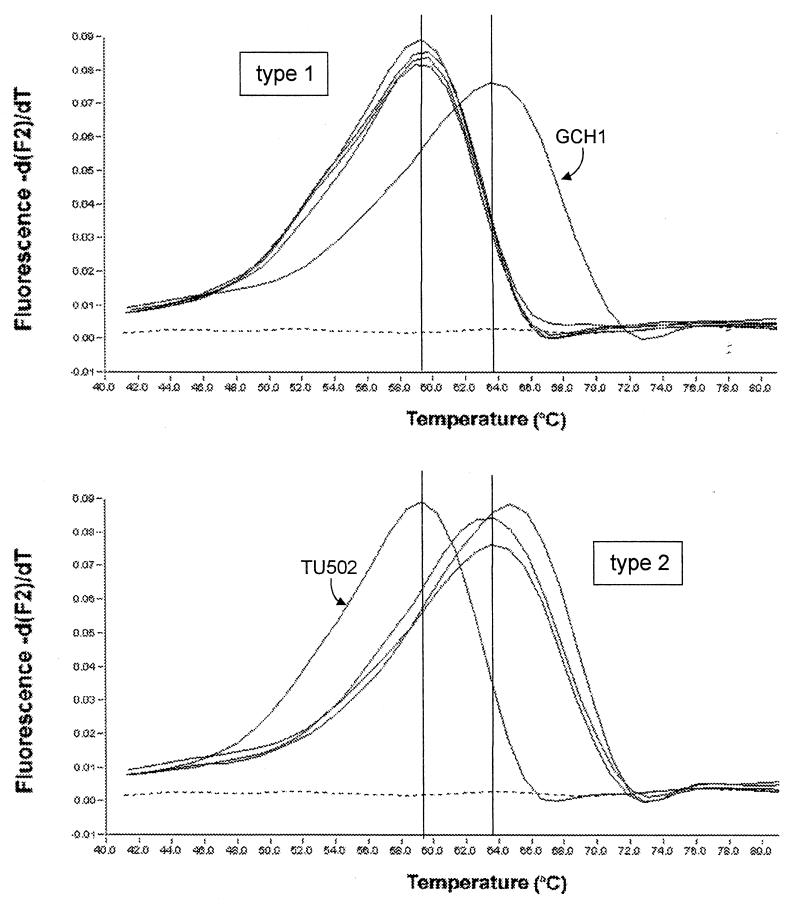
FIG. 2.
Real-time PCR genotyping of C. parvum with two fluorescent probes specific for the β-tubulin sequence. Shown is the MCA for four type 1 isolates and three type 2 isolates. The cursors were manually positioned on the melting peaks of reference isolates GCH1 and TU502. The melting curves for type 1 isolates TU502, UG1544, UG1259, and UG1610 and type 2 isolates GCH1 (32), TAMU (25), and OHIO are shown.
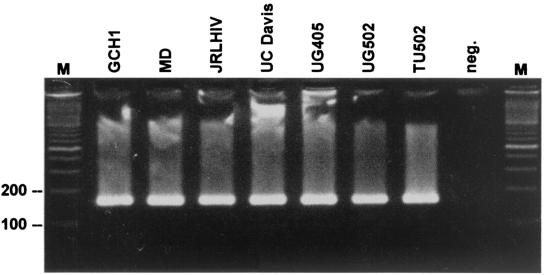
FIG. 3.
Gel analysis of amplicons from different C. parvum isolates obtained by PCR with primers btub1 and btub2. Amplicons were fractionated on 1.5% agarose. Isolate designations are shown at the top. Lane M, 100-bp ladder. The positions of the 100- and 200-bp markers are indicated.
Differentiation between C. parvum type 1 and type 2 by SYBR Green I real-time PCR.
To assess the feasibility of using a previously described polymorphic coding sequence (5) for genotyping of C. parvum by SYBR Green I MCA, a 319-bp fragment from the GP900/poly(T) locus was amplified from seven type 2 and four type 1 C. parvum isolates. The amplicon is located within the gene encoding the GP900 sporozoite surface antigen (3) and was previously shown to discriminate between type 1 and type 2 isolates on the basis of a polymorphic RsaI restriction site (5). MCA revealed different melting temperatures for type 1 and type 2 sequences (Fig. 4), although the difference in melting temperatures between types was smaller than that observed by the probe-based assay described above. The mean melting temperature for three type 1 isolates was 83.47°C (SD = 0.06), whereas for seven type 2 isolates the average melting temperature was 83.04°C (SD = 0.12). Statistical analysis demonstrated that this difference was also significant (t = 5.73; P < 0.001). The sequence polymorphisms responsible for different melting peak temperatures were investigated by cloning and sequencing of the poly(T) amplicon from two type 2 isolates (isolates GCH1 and JRLHIV) and one type 1 isolate (isolate TU502). Nine SNPs between type 1 and type 2 were identified within the 319-bp amplicon, as was a CCA deletion at position 1542 (GenBank accession no. U83169) in the type 2 sequences. Of the nine SNPs, four were A or T→G or C changes, four were G or C→A or T changes, and one was a T (type 2)→A (type 1) change. Consistent with the tight distribution of the type 2 melting peak temperatures, the two type 2 sequences were identical. Since the observed SNPs do not change the number of A or T and G or C residues, we conclude that the difference in the melting temperature between the two types is caused by the 3-bp deletion in type 2.
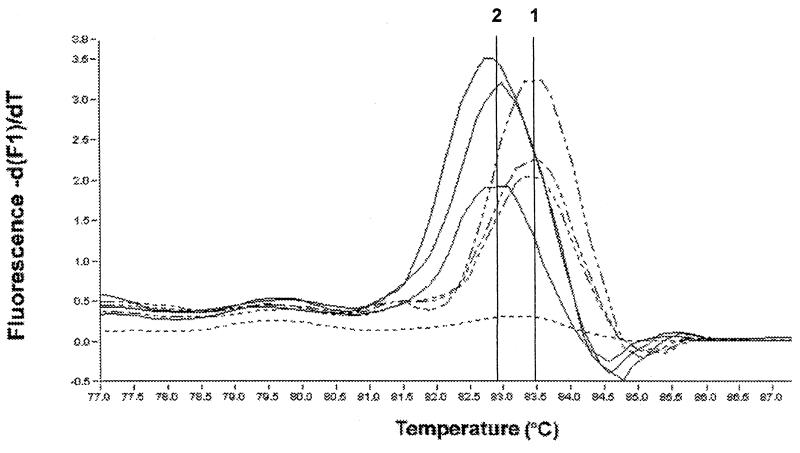
FIG. 4.
Real-time PCR genotyping of C. parvum using SYBR Green I fluorescence. A polymorphic 319-bp fragment of the GP900 gene (2) was amplified by PCR in the presence of SYBR Green I. Shown are the melting peaks for type 1 isolates 2066-K (37), TU502, and CDC728 and type 2 isolates 7C (10), TAMU (25), and JRLHIV. The differences in melting peak temperatures between the type 1 and the type 2 isolates were statistically significant at the 0.001 level. The cursors were positioned manually and are labeled with the respective C. parvum type.
DISCUSSION
Several real-time PCR methods for detection and genotyping of C. parvum oocysts were developed. As with conventional PCR, the sensitivity of the real-time method was strongly dependent on the amplification primers, even when primer pairs amplifying overlapping regions of the same gene were compared. Even though the sequence detected with primers btub1 and btub2 is present at only a single copy (26), single oocysts isolated by micromanipulation were detected in more than 50% of the PCR tests. The sensitivity of the method in which oocysts were introduced directly into the PCR mixture appeared to be equal to those of methods in which DNA was extracted prior to PCR. Two series of negative control experiments with washed and unwashed oocyst suspensions were performed to investigate the origin of the C. parvum DNA detected in some of the negative control reactions and specifically to assess whether additional oocysts may accidentally have been transferred to the PCR mixture during the micromanipulation procedure or whether DNA was present in solution. The latter possibility was consistent with the fact that the oocysts used in these experiments had been stored in the same suspension at 4°C for approximately 3 months. During storage, a fraction of oocysts could have excysted and DNA from degrading sporozoites could have been released into solution. The absence of positive reactions for the second series of negative controls with a newly washed oocyst suspension was consistent with this assumption and ruled out the possibility of accidental transfer of oocysts during micromanipulation.
The development of real-time PCR methods capable of discriminating between C. parvum type 1 and type 2 and other host-associated genotypes is relevant to the study of the epidemiology of this parasite and for source tracking. Type 1 and type 2 are thought to be transmitted by different routes since they differ in their host specificities. Type 1 has been detected almost exclusively in human infections, whereas type 2 infects both humans and a variety of animal species (2, 33). The melting temperatures observed with fluorescent probes and with SYBR Green I were highly reproducible, indicating that these methods are suitable for differentiating C. parvum types, even on the basis of SNPs or small nucleotide deletions. In 11 independent experiments the coefficient of variation (CV) for the melting temperature of isolate TU502 was only 1.4%, and the CV was 3% for 10 replicate experiments with GCH1. CVs for the melting temperature of the poly(T) amplicon observed with SYBR Green I were as low as 0.1%. A possible source of the relatively small variability in melting temperature is the template concentration. As visible in Fig. 1, higher oocyst concentrations appeared to correlate with slightly lower melting temperatures, a phenomenon also observed in serial dilution experiments with larger oocyst numbers.
For genotyping, assays based on fluorescent probes have higher specificities, but the requirement for primers with a fluorescent label increases the cost. The results of our unpublished experiments with type 1 and type 2 DNA mixed in different ratios indicate that probes do not resolve mixtures of templates as well as the SYBR Green I method does. This limitation is a concern in epidemiological studies, in which it is of interest to detect genotypically mixed parasite populations. In contrast, preliminary experiments with cloned microsatellite alleles of C. parvum mixed at different ratios showed that the limit of detection for a minority template was approximately 10%. This compared favorably with the 25% limit that we previously observed by restriction fragment length polymorphism analysis and is similar to the results recently reported by Reed et al. (27).
The present laboratory methods for the diagnosis of C. parvum by microscopy are generally adequate for samples with high concentrations of oocysts, but are insufficient for the detection of cases of cryptosporidiosis in which only small numbers of oocysts are excreted (6). The high sensitivity of real-time PCR will facilitate the detection of asymptomatic carriers. Such information could be valuable for epidemiological studies of human and animal hosts. The time savings of the present approach are considerable. With the real-time PCR cycler used in these experiments, a 40-cycle amplification was completed in only 35 to 45 min. The fact that no gel analysis is required for routine experiments reduces the time needed by at least 1 h and could facilitate the implementation of high-throughput screening. The fact that the entire amplification and sample analysis are performed in a sealed capillary tube also reduces the risk of contamination from DNA carryover.
The full potential of the real-time method for the detection of diagnostic DNA sequences will be achieved only with the simultaneous detection of different species of pathogens by using the capability of most real-time PCR machines to measure fluorescence at different wavelengths. For instance, one could envision an assay in which multiple species of a pathogen are detected with SYBR Green I on the F1 channel and the genotype of one of these species is identified on the F2 channel with an internal probe. In the case of Cryptosporidium, a possible setup would be to identify the species present in water or fecal samples by SYBR Green I MCA and the C. parvum genotype with fluorescent probes.
Acknowledgments
Financial support from the Çukurova University Research Fund (grant TF-2000.M.10) and the U.S. Department of Agriculture National Research Initiative (grant 9800919) is gratefully acknowledged.
Our thanks go to Kerem Ozgunen, Çukurova University, for help with oocyst micromanipulations; to Olfert Landt, TIB Molbiol, for designing the probes; and to James K. Tumwine, Addy Kekitiinwa, Nicolette Nabukeera, Cynthia Chappell, Lucy Ward, Pablo Okhuysen, and Edward Atwill for oocysts.
REFERENCES
- 1.Aarnaes, S. L., J. Blanding, S. Speir, D. Fortal, L. M. de la Maza, and E. M. Petersen. 1994. Comparison of ProSpecT and Color Vue enzyme-linked immunoassays for the detection of Cryptosporidium in stool specimens. Diagn. Microbiol. Infect. Dis. 19:221-225. [DOI] [PubMed] [Google Scholar]
- 2.Awad-El-Kariem, F. M., H. A. Robinson, F. Petry, V. McDonald, D. Evans, and D. Casemore. 1998. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol. Res. 84:297-301. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, D. A., A. Bonnin, J. X. Huang, L. Gousset, J. Wu, J. Gut, P. Doyle, J. F. Dubremetz, H. Ward, and C. Petersen. 1998. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol. Biochem. Parasitol. 96:93-110. [DOI] [PubMed] [Google Scholar]
- 4.Cacciò, S., W. Homan, K. van Dijk, and E. Pozio. 1999. Genetic polymorphism at the beta-tubulin locus among human and animal isolates of Cryptosporidium parvum. FEMS Microbiol. Lett. 170:173-179. [DOI] [PubMed] [Google Scholar]
- 5.Carraway, M., S. Tzipori, and G. Widmer. 1997. New RFLP marker in Cryptosporidium parvum identifies mixed parasite populations and genotypic instability in response to host change. Infect. Immun. 65:3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell, C. L., P. C. Okhuysen, C. R. Sterling, and H. L. DuPont. 1996. Cryptosporidium parvum: intensity of infection and oocyst excretion patterns in healthy volunteers. J. Infect. Dis. 173:232-236. [DOI] [PubMed] [Google Scholar]
- 7.Current, W. L., and L. S. Garcia. 1991. Cryptosporidiosis. Clin. Microbiol. Rev. 4:325-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, M. Q., D. O. Cliver, and T. W. Mariam. 1997. Immunomagnetic capture PCR to detect viable Cryptosporidium parvum oocysts from environmental samples. Appl. Environ. Microbiol. 63:3134-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Giovanni, G. D., F. H. Hashemi, N. J. Shaw, F. A. Abrams, M. W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl. Environ. Microbiol. 65:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, X., S. M. Rich, S. Tzipori, and G. Widmer. 2002. Experimental evidence for genetic recombination in the opportunistic pathogen Cryptosporidium parvum. Mol. Biochem. Parasitol. 119:55-62. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, L. S., A. C. Shum, and D. A. Bruckner. 1992. Evaluation of a new monoclonal antibody combination reagent for direct fluorescence detection of Giardia cysts and Cryptosporidium oocysts in human fecal specimens. J. Clin. Microbiol. 30:3255-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasser, R. B., X. Zhu, S. Caccio, R. Chalmers, G. Widmer, U. M. Morgan, R. C. Thompson, E. Pozio, and G. F. Browning. 2001. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22:433-437. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths, J. K. 1998. Human cryptosporidiosis: epidemiology, transmission, clinical disease, treatment, and diagnosis. Adv. Parasitol. 40:38-87. [DOI] [PubMed] [Google Scholar]
- 14.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, J. A., R. Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323-337. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., F. Constantini, and E. Lacy. 1986. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 6:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger, P., A. Wiedenmann, D. Tougianidou, and K. Botzenhart. 2001. Quantitative detection of Cryptosporidium parvum after in vitro excystation by LightCycler PCR, p. 341-348. In S. Meyer, C. Witter, and K. I. Nakagawara (ed.), Rapid real-time PCR methods and applications. Springer-Verlag Berlin, Germany.
- 19.Lally, N. C., G. D. Baird, S. J. McQuay, F. Wright, and J. J. Oliver. 1992. A 2359-base pair DNA fragment from Cryptosporidium parvum encoding a repetitive oocyst protein. Mol. Biochem. Parasitol. 56:69-78. [DOI] [PubMed] [Google Scholar]
- 20.Laxer, M. A., B. D. Timblin, and R. J. Patel. 1991. DNA sequences for specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 45:688-694. [DOI] [PubMed] [Google Scholar]
- 21.Ma, P., and R. Soave. 1983. Three-step stool examination for cryptosporidiosis in 10 homosexual men with protracted watery diarrhea. J. Infect. Dis. 147:824-828. [DOI] [PubMed] [Google Scholar]
- 22.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan, U. M., C. C. Constantine, P. O'Donoghue, B. P. Meloni, P. A. O'Brien, and R. C. A. Thompson. 1995. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am. J. Trop. Med. Hyg. 52:559-564. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, U. M., P. T. Monis, R. Fayer, P. Deplazes, and R. C. Thompson. 1999. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J. Parasitol. 85:1126-1133. [PubMed] [Google Scholar]
- 25.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 26.Piper, M. B., A. T. Bankier, and P. H. Dear. 1998. A HAPPY map of Cryptosporidium parvum. Genome Res. 8:1299-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, C., G. D. Sturbaum, P. J. Hoover, and C. R. Sterling. 2002. Cryptosporidium parvum mixed genotypes detected by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 68:427-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 29.Rochelle, P. A., R. De Leon, A. Johnson, M. H. Stewart, and R. L. Wolfe. 1999. Evaluation of immunomagnetic separation for recovery of infectious Cryptosporidium parvum oocysts from environmental samples. Appl. Environ. Microbiol. 65:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulaiman, I. M., A. A. Lal, M. J. Arrowood, and L. Xiao. 1999. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J. Parasitol. 85:154-157. [PubMed] [Google Scholar]
- 31.Sulaiman, I. M., L. Xiao, and A. A. Lal. 1999. Evaluation of Cryptosporidium parvum genotyping techniques. Appl. Environ. Microbiol. 65:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzipori, S., W. Rand, J. K. Griffiths, G. Widmer, and J. Crabb. 1994. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin. Diagn. Lab. Immunol. 1:450-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzipori, S., and G. Widmer. 2000. The biology of Cryptosporidium. Contrib. Microbiol. 6:1-32. [DOI] [PubMed] [Google Scholar]
- 34.Valdez, L. M., H. Dang, P. C. Okhuysen, and C. L. Chappell. 1997. Flow cytometric detection of Cryptosporidium oocysts in human stool samples. J. Clin. Microbiol. 35:2013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widmer, G. 1998. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv. Parasitol. 40:224-241. [DOI] [PubMed] [Google Scholar]
- 36.Widmer, G., L. Tchack, C. L. Chappell, and S. Tzipori. 1998. Sequence polymorphism in the β-tubulin gene reveals heterogeneous and variable population structure in Cryptosporidium parvum. Appl. Environ. Microbiol. 64:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]