Abstract
The sodium dodecyl sulfate-polyacrylamide gel electrophoresis patterns of water-soluble proteins and the randomly amplified polymorphic DNA (RAPD) patterns of whole-cell lysates from 21 Aspergillus ustus isolates, including 11 reference strains and 10 patient and environmental strains from one hospital, were investigated. All isolates showed identical protein patterns. The RAPD assay discriminated between all reference strains. Comparison of hospital isolates showed identical RAPD patterns in some of the patient and environmental isolates. The data indicate that the RAPD technique is useful for fingerprinting A. ustus.
Aspergillus ustus is a rare cause of infections in humans. To date, only nine cases have been reported (5, 14). However, most cases were published after 1990, which indicates that this species might be an emerging pathogen. A large number of studies of the molecular epidemiology of Aspergillus fumigatus have been published (for a review, see reference 7). In contrast, data on the phenotypical and genotypical characteristics of other Aspergillus spp. are scant. Recently, a study of Aspergillus terreus infections in immunosuppressed patients showed that environmental and some patient isolates had identical band patterns by randomly amplified polymorphic DNA (RAPD) analysis (6). However, fingerprinting techniques to study A. ustus have not been established. Therefore, we chose to study the usefulness of two fingerprinting techniques (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] and the RAPD assay) for analyzing A. ustus isolates.
Eleven reference strains (Table 1) were obtained from international culture collections (Centraalbureau voor Schimmelcultures [CBS], Utrecht, The Netherlands; Biomedical Fungi and Yeast Collection [IHEM], Brussels, Belgium; National Collection of Pathogenic Fungi [NCPF], Bristol, United Kingdom). The remaining 10 isolates (AZN) were from the University Medical Center, Nijmegen, The Netherlands. A detailed description of these isolates and their origins (Table 1, isolates 12 to 21) is given elsewhere (14). An analysis of the protein patterns of water-soluble proteins of whole-cell lysates in Coomassie blue-stained SDS-PAGE gels (7.5%) was performed as described previously (11).
TABLE 1.
Sources and RAPD types of 21 A. ustus isolates
| Isolatea | Designation | Source (yr of isolation) | Type of RAPD pattern produced by primerb:
|
Combined typec | ||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | ||||
| 1 | CBS 596.65 | Sugar; Louisiana (1965) | A-1 | B-1 | C-1 | D-1 | E-1 | A/D-1 |
| 3 | IHEM 1139 | Air; Brussels, Belgium (1982) | A-2 | B-2 | C-2 | D-2 | E-2 | A/D-2 |
| 4 | IHEM 9696 | Indoor air (hospital); Marseille, France (1991) | A-3 | B-3 | C-3 | D-3 | E-3 | A/D-3 |
| 5 | CBS 209.92 | Soils; La Palma, Spain (1992) | A-4 | B-4 | C-4 | D-4 | E-3 | A/D-4 |
| 6 | CBS 133.55 | Textile buried in soil; Utrecht, The Netherlands (1955) | A-5 | B-5 | C-5 | D-5 | E-4 | A/D-5 |
| 2 | CBS 102278 | Human, subcutaneous infection; Utrecht, The Netherlands (1999) | A-6 | B-6 | C-6 | D-6 | E-5 | A/D-6 |
| 7 | NCPF 2848 | Human, brain; Bristol, United Kingdom (1991) | A-3 | B-3 | C-3 | D-7 | E-3 | A/D-7 |
| 8 | NCPF 2951 | Human, sputum; Bristol, United Kingdom (1993) | A-3 | B-3 | C-3 | D-8 | E-3 | A/D-8 |
| 9 | NCPF 2922 | Human, ethmoid sinus; Bristol, United Kingdom (1993) | A-7 | B-3 | C-3 | D-3 | E-3 | A/D-9 |
| 10 | NCPF 2744 | Human, sputum; Bristol, United Kingdom (1990) | A-8 | B-7 | C-3 | D-9 | E-6 | A/D-10 |
| 11 | NCPF 2766 | Human, mitral valve prosthesis; Bristol, United Kingdom (1990) | A-9 | B-8 | C-3 | D-10 | E-3 | A/D-11 |
| 12 | AZN 677 | Bronchoalveolar lavage specimen, patient 1; University Medical Center, Nijmegen, The Netherlands (1992) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
| 13 | AZN 678 | Sputum, patient 1; University Medical Center, Nijmegen, The Netherlands (1992) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
| 14 | AZN 682 | Autopsy specimen, patient 1; University Medical Center, Nijmegen, The Netherlands (1992) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
| 15 | AZN 741 | Hematology ward, room; University Medical Center, Nijmegen, The Netherlands (1993) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
| 16 | AZN 924 | Hematology ward, room; University Medical Center, Nijmegen, The Netherlands (1993) | A-11 | B-10 | C-3 | D-12 | E-3 | A/D-13 |
| 17 | AZN 943 | Laboratory contaminant; University Medical Center, Nijmegen, The Netherlands (1993) | A-12 | B-10 | C-8 | D-13 | E-3 | A/D-14 |
| 18 | AZN 2725 | Sputum, patient 2; University Medical Center, Nijmegen, The Netherlands (1995) | A-13 | B-10 | C-8 | D-12 | E-3 | A/D-15 |
| 19 | AZN 3297 | Feces, patient 3; University Medical Center, Nijmegen, The Netherlands (1995) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
| 20 | AZN 6989 | Ascites fluid, patient 4; University Medical Center, Nijmegen, The Netherlands (1997) | A-11 | B-10 | C-3 | D-12 | E-3 | A/D-13 |
| 21 | AZN 7134 | Hematology ward, room; University Medical Center, Nijmegen, The Netherlands (1997) | A-10 | B-9 | C-7 | D-11 | E-3 | A/D-12 |
For a detailed description of isolates 12 to 21, see reference 14.
Numbers of different RAPD types for the following primers: A, 13; B, 10; C, 8; D, 13; E, 6.
Results of the combined analysis of the RAPD patterns produced by primers A and D. The number of different RAPD types for the combined type was 15.
For the RAPD assay, extraction of DNA was performed as previously described (12). Briefly, mycelial cells (1 g [wet weight]) from 24-h-old broth cultures were disrupted by using glass beads and DNA was isolated by using phenol-chloroform, ammonium acetate, isopropanol, and RNase A.
Amplification was carried out with four single primers that have already been successfully used with other Aspergillus spp. (6, 10-12): primer A, 5′-GTA TTG CCC T-3′ (1); primer B, 5′-GCT GGT GG-3′ (8); primer C, 5′-TCA CCC TGG A-3′ (12); and primer D, 5′-(GATA)4-3′ (13). The primers were run under the same conditions (master mix containing 3 mM MgCl2, 200 pmol of each primer, and 5 ng of DNA; final volume, 100 μl; 45 cycles at 94°C for 1 min, 35°C for 2 min, and 72°C for 2 min in a Minicycler [MJ Research, Watertown, Mass.]). For primer E (5′-GAG GGT GGC GGT TCT-3′), which was used to fingerprint Pseudallescheria boydii and yeasts (9, 15), 4 mM MgCl2 and an annealing temperature of 50°C were used. After electrophoretic separation in 1.6% agarose gels, the ethidium bromide-stained band patterns were analyzed by using Imagemaster 1D Elite software (Amersham Pharmacia Biotech, Freiburg, Germany). The similarity of band patterns was estimated by means of the Jaccard comparison, and the clustering was determined by the unweighted-pair-group method with averages. Band patterns of >90% similarity were classified as identical (3). Reproducibility of band patterns was demonstrated by analysis of two to three different subcultures of each strain.
By SDS-PAGE, all 21 A. ustus isolates showed numerous bands in the range of 115 to 20 kDa. Major bands were seen at 61, 57 to 55, 53, 47, 43, and 37 kDa (data not shown) but the patterns of the isolates did not differ. This method is obviously not useful for fingerprinting A. ustus. These same results were found for A. fumigatus, Aspergillus niger (10; our unpublished results), and A. terreus (11). In Aspergillus flavus and Aspergillus (Emericella) nidulans, however, different protein patterns were found within the species (10). The basis for the different phenotypic features of the species of Aspergillus is not known but may indicate genetic recombination or different subspecies.
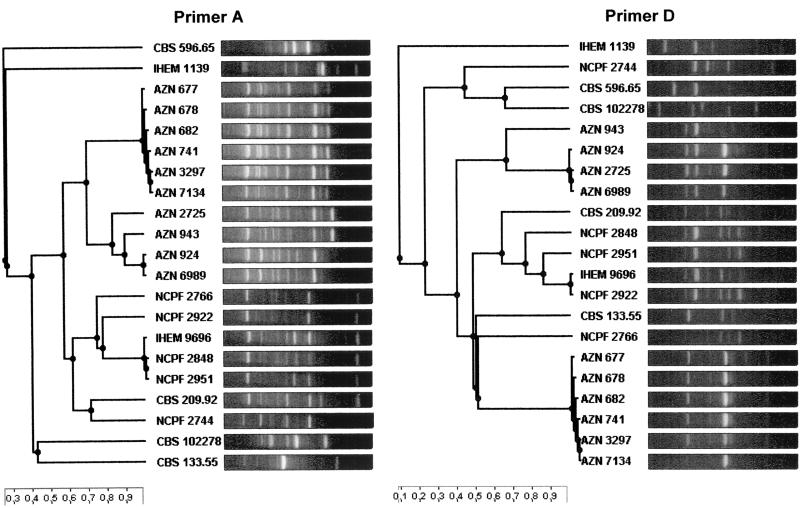
In contrast to the phenotypic method, the RAPD analysis showed a high degree of discriminatory power. Amplification with primers A and D resulted in 13 different RAPD types of patterns each, whereas amplification with primers B, C, and E yielded 10, 8, and 6 different patterns, respectively. In Fig. 1, the cluster analyses of the band patterns of the 21 strains produced by primers A and D are depicted. The combined analyses of the patterns of these two primers resulted in complete discrimination of the 11 reference strains (Table 1, combined types A/D-1 to A/D-11). When investigating the isolates from patients and the environment from the hospital in Nijmegen, The Netherlands (isolates 12 to 21), we found that primer A showed the highest degree of discriminatory power with four different types (A-10 to A-13). Combined analysis of patterns produced by different primers resulted in no further discrimination (A/D-12 to A/D-15). As expected, all three of the isolates obtained from one patient (Table 1, isolates 12 to 14) showed identical patterns (RAPD combined type A/D-12). However, the same pattern was found in two environmental isolates obtained from the ward (isolates 15 and 21) and in one isolate obtained from a patient (isolate 19) from a different ward. Furthermore, two other isolates (isolate 16 from the environment and isolate 20 from a patient) showed an identical RAPD type (combined type A/D-13). This finding indicates that at least some RAPD combined types (A/D-12 and A/D-13) persisted over long periods (up to 6 years) in the environment. Similar results were found for A. fumigatus (4) and A. terreus (6).
FIG. 1.
Cluster analysis of RAPD patterns of 21 A. ustus strains (11 reference strains and 10 patient and environmental isolates from Nijmegen, The Netherlands) produced by primers A and D. The scales at the bottom represent the similarity index. AZN, isolates from Nijmegen, The Netherlands; CBS, NCPF, and IHEM, reference strains from culture collections.
The present data verify that RAPD analysis is useful for fingerprinting not only A. fumigatus (1, 7, 8, 10, 12), A. flavus (10), A. nidulans (10), and A. terreus (6, 11) but also A. ustus when the same primers (A and D) are used. Primer E (core sequence of phage M13), which was successfully used for fingerpinting P. boydii (9) and yeasts (15), showed a minor degree of discriminatory power in the present study.
Since other published techniques for fingerprinting aspergilli, e.g., the microsatellite PCR described by Bart-Delabesse et al. (2) or the Southern hybridization probe described by Debeaupuis et al. (4), work only with A. fumigatus (4; our unpublished results), the RAPD technique is at present the sole method which is generally applicable for the typing of various Aspergillus spp., including A. ustus.
Acknowledgments
We thank Dirk Schmidt for excellent technical assistance.
This study was sponsored in part by a grant from the University Hospital Essen (IFORES).
REFERENCES
- 1.Aufauvre-Brown, A., J. Cohen, and D. W. Holden. 1992. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J. Clin. Microbiol. 30:2991-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bart-Delabesse, E., J.-F. Humbert, É. Delabesse, and S. Bretagne. 1997. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollet, C., and P. de Micco. 1992. Taxonomic methods, p. 179-200. In J. Lederberg (ed.), Encyclopedia of microbiology, vol 4. Academic Press, San Diego, Calif.
- 4.Debeaupuis, J. P., J. Sarfati, V. Chazalet, and J. P. Latgé. 1997. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 65:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gené, J., A. Azón-Masoliver, J. Guarro, G. de Febrer, A. Martinez, C. Grau, M. Ortoneda, and F. Ballester. 2001. Cutaneous infection caused by Aspergillus ustus, an emerging opportunistic fungus in immunosuppressed patients. J. Clin. Microbiol. 39:1134-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lass-Flörl, C., P.-M. Rath, D. Niederwieser, G. Kofler, R. Würzner, A. Krezy, and M. P. Dierich. 2000. Aspergillus terreus infections in haematological malignancies: molecular epidemiology suggests association with in-hospital plants. J. Hosp. Infect. 46:31-35. [DOI] [PubMed] [Google Scholar]
- 7.Latgé, J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loudon, K. W., J. P. Burnie, A. P. Coke, and R. C. Matthews. 1993. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of polymorphic DNA. J. Clin. Microbiol. 31:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rainer, J., G. S. de Hoog, M. Wedde, Y. Gräser, and S. Gilges. 2000. Molecular variability of Pseudallescheria boydii, a neurotropic opportunist. J. Clin. Microbiol. 38:3267-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rath, P.-M. 2001. Phenotypic and genotypic characterization of reference strains of the genus Aspergillus. Mycoses 44:65-72. [DOI] [PubMed] [Google Scholar]
- 11.Rath, P.-M., S. Kamphoff, and R. Ansorg. 1999. Value of different methods for the characterisation of Aspergillus terreus strains. J. Med. Microbiol. 48:161-166. [DOI] [PubMed] [Google Scholar]
- 12.Rath, P.-M., G. Marggraf, H. Dermoumi, and R. Ansorg. 1995. Use of phenotypic and genotypic fingerprinting methods in the strain identification of Aspergillus fumigatus. Mycoses 38:429-434. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan, D., D. Bennett, M. Henman, P. Harwood, S. Flint, F. Mulcahy, D. Shanley, and D. Coleman. 1993. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J. Clin. Microbiol. 31:2124-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verweij, P. E., M. F. Q. van den Bergh, P. M. Rath, B. E. De Pauw, A. Voss, and J. F. G. M. Meis. 1999. Invasive aspergillosis caused by Aspergillus ustus: case report and review. J. Clin. Microbiol. 37:1606-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu, J., C. M. Boyd, E. Livingston, W. Meyer, J. F. Madden, and T. G. Mitchell. 1999. Species and genotypic diversities and similarities of pathogenic yeasts colonizing women. J. Clin. Microbiol. 37:3835-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]