Abstract
Usually a saprophyte, Scedosporium apiospermum often colonizes the respiratory tracts of patients with cystic fibrosis (CF). In order to improve our understanding of the molecular epidemiology of the airway colonization, 129 sequential and multiple isolates collected from January 1998 to March 1999 from nine CF patients monitored in three hospitals in France were typed by random amplification of polymorphic DNA with primers GC70, UBC-701, and UBC-703. Among these primers, UBC-703 was the most discriminating, allowing the differentiation of 14 genotypes. Combining the results obtained with this three-primer set resulted in the differentiation of 16 genotypes. No common genotype was found among the different patients, and no clustering according to geographic origin of the isolates was seen. In addition, five of the patients were colonized by a single genotype. The others usually exhibited a predominant genotype accompanied by one or two others, which were found occasionally and were genetically close to the predominant genotype. Thus, our study demonstrates the persistence of the fungus despite antifungal treatments and therefore reinforces the need for the development of new antifungals that are more efficient against this species.
Cystic fibrosis (CF) is the most common inherited disease in the Caucasian population; it results from mutations in the gene CFTR, which encodes the chloride channel CF transmembrane regulator, and is characterized by a dysfunction of the exocrine glands. As a consequence of the resulting abnormal viscosity of the bronchial secretions, the lungs of patients with CF are often colonized by or infected with various bacteria, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Burkholderia cepacia (9). At a more advanced stage of the disease, the respiratory tracts of these patients may also be colonized by filamentous fungi because of the entrapment of airborne mold spores in the viscous mucus and because of the bronchopulmonary epithelial tissue damage induced by bacterial proteases and leukocyte elastase. Besides Aspergillus fumigatus, which remains by far the most common species, other filamentous fungi may also be encountered. In a large prospective study encompassing 128 CF patients monitored during a 5-year period, Scedosporium apiospermum was isolated from bronchial secretions from 8.6% of the patients, ranking it as the second most frequently occurring filamentous fungus identified (3).
Usually a saprophyte growing in polluted soils and sewage, this filamentous fungus may cause in humans various mycoses resulting from traumatic or iatrogenic inoculation of telluric conidia, including white-grain mycetoma, keratitis, endophthalmitis, osteomyelitis, and endocarditis (15, 21, 22). Respiratory diseases may also occur in receptive hosts, likely due to the inhalation of airborne conidia, and are similar to those described for A. fumigatus, i.e., unilateral sinusitis, necrotizing pneumonia, fungus ball in the lung, and allergic bronchopulmonary mycosis, as well as disseminated infections in severely immunocompromised hosts. More recently, attention has been paid to the particular occurrence of this fungus in CF patients (2, 3). However, its pathogenic role in this context remains controversial. In addition, little information is available concerning the epidemiology of the colonization of the airways by this fungus. In order to improve our knowledge in this field, sequential and multiple isolates from different CF patients were typed by random amplification of polymorphic DNA (RAPD) with a three-primer set previously selected for its ability to discriminate among epidemiologically unrelated strains of S. apiospermum (26).
MATERIALS AND METHODS
Patients and isolates.
From January 1998 to March 1999, all CF patients monitored in the pediatric departments of hospitals in Angers, Giens, and Tours (France) were surveyed for colonization of the respiratory tract by S. apiospermum. To this end, sputum samples collected during the routine annual clinical visit of the patients or during an admission to the hospital because of exacerbation of their pulmonary disease were inoculated on yeast extract-peptone-dextrose agar plates, which contained the following: yeast extract, 5 g; peptone, 10 g; glucose, 20 g; chloramphenicol 1.0 g; agar, 20 g; and distilled water, 1 liter. Since S. apiospermum is often associated with A. fumigatus in CF patients, the culture medium was supplemented with cycloheximide (1.0 g per liter) as previously described (2). This concentration of cycloheximide inhibits the growth of most strains of A. fumigatus, whereas S. apiospermum tolerates cycloheximide concentrations up to 5 g per liter (6). Aliquots (100 μl) of the samples were plated directly or after digestion for 30 min at 37°C with an equal volume of 2,3-dihydroxy-1,4-dithiolbutane (Digest-EUR; Eurobio, Les Ulis, France), and this was followed by 1:10 dilution in sterile distilled water (2). Two plates were used for each sample, and after incubation at 37°C, all colonies (with a maximum of five colonies for each sample) from the primary cultures were isolated before sporulation. All isolates were preserved and referenced at the BCCM/IHEM Culture Collection (Brussels, Belgium). The origin of the isolates and patient clinical data (dates of birth, of CF diagnosis, and of first colonization of the airways by S. apiospermum, as well as dates of antifungal treatments) are indicated in Tables 1 and 2. Colonization of the airways by S. apiospermum was defined by the isolation of the fungus from at least three successive sputum samples collected during a 6-month period. Bronchopulmonary infection was defined by clinical or radiological signs associated with positive sputum cultures and with serum specific antibodies revealed by counterimmunoelectrophoresis using both culture filtrate and a somatic extract of S. apiospermum (3).
TABLE 1.
Clinical data from CF patients colonized by S. apiospermum
| Patient no. (hospital location) | Date of birth (mo/day/yr) | Gendera | Yr of CF diagnosis (mutation) | Date of first colonization by S. apiospermum | Patient status regarding S. apiospermum | Antifungal therapy during the study period |
|---|---|---|---|---|---|---|
| P1 (Angers) | 12/20/1979 | M | 1984 (ΔF508/ΔF508) | December 1991 | Bronchial colonization | Itraconazole from June 1998 to March 1999 |
| P2 (Angers) | 4/6/1978 | M | 1984 (ΔF508/ΔF508) | November 1993 | Transient carriage | None |
| P3 (Giens) | 3/15/1972 | F | 1973 (ΔF508/ΔF508) | March 1989 | Bronchial colonization | Itraconazole continuously during this period |
| P4 (Giens) | 12/15/1950 | M | 1996 (ΔF508/2789 + 5G → A) | January 1997 | Bronchial colonization | Itraconazole continuously during this period |
| P5 (Giens) | 4/12/1987 | M | 1994 (unknown) | September 1998 | Transient carriage | Itraconazole from September to November 1998 |
| P6 (Giens) | 12/17/1984 | F | 1985 (G542X/unknown) | January 1992 | Bronchial colonization | Itraconazole from October to December 1998 |
| P7 (Giens) | 5/21/1977 | F | 1990 (ΔF508/S1251N) | February 1994 | Bronchial colonization | Itraconazole from January 1998 to March 1998 and July 1998 to September 1998 |
| P8 (Tours) | 3/14/1978 | F | 1988 (ΔF508/ΔF508) | December 1993 | Bronchopulmonary infection | Fluconazole until April 1998, itraconazole from March 1998 to February 1999, voriconazole since March 1999 |
| P9 (Tours) | 10/25/1984 | F | 1988 (ΔF508/ΔF508) | August 1995 | Bronchial colonization | None |
Abbreviations: M, male; F, female.
TABLE 2.
Genotypes of S. apiospermum isolates from CF patients
| Patient no. | Location of hospital | Isolation date (mo/day/yr) | IHEMb isolate no. | Electrophoretic patterna obtained using primer:
|
Overall genotype | ||
|---|---|---|---|---|---|---|---|
| GC70 | UBC-701 | UBC-703 | |||||
| P1 | Angers | 1/9/1998 | 14263 | A | A | A | 1 |
| 14264 | A | A | A | 1 | |||
| 14265 | A | A | A | 1 | |||
| 14266 | A | A | A | 1 | |||
| 14267 | A | A | A | 1 | |||
| 2/16/1998 | 14368 | A | A | A | 1 | ||
| 14369 | A | A | A | 1 | |||
| 14370 | A | A | A | 1 | |||
| 2/18/1998 | 14363 | A | A | A | 1 | ||
| 14364 | A | A | A | 1 | |||
| 14366 | A | A | A | 1 | |||
| 3/10/1998 | 14457 | A | A | A | 1 | ||
| 14458 | A | A | A | 1 | |||
| 14459 | A | A | A | 1 | |||
| 14460 | A | A | A | 1 | |||
| 14461 | A | A | A | 1 | |||
| 4/17/1998 | 15546 | A | A | A | 1 | ||
| 15547 | A | A | A | 1 | |||
| 15548 | A | A | A | 1 | |||
| 15549 | A | A | A | 1 | |||
| 6/10/1998 | 14638 | B | A | A | 2 | ||
| 14639 | A | A | A | 1 | |||
| 14640 | A | A | A | 1 | |||
| 14641 | A | A | A | 1 | |||
| 14642 | A | A | A | 1 | |||
| 7/31/1998 | 14756 | A | A | A | 1 | ||
| 14757 | A | A | A | 1 | |||
| 14758 | C | B | B | 3 | |||
| 14759 | A | A | A | 1 | |||
| 14760 | A | A | A | 1 | |||
| 9/4/1998 | 15139 | A | A | A | 1 | ||
| 15140 | A | A | A | 1 | |||
| 15141 | A | A | A | 1 | |||
| 14142 | A | A | A | 1 | |||
| 14143 | A | A | A | 1 | |||
| 1/13/1999 | 15581 | A | A | A | 1 | ||
| 15582 | A | A | A | 1 | |||
| 15583 | A | A | A | 1 | |||
| P2 | Angers | 1/13/1999 | 15579 | D | C | C | 4 |
| 15580 | D | C | C | 4 | |||
| P3 | Giens | 2/24/1998 | 14451 | E | D | D | 5 |
| 14452 | E | D | D | 5 | |||
| 14453 | E | D | D | 5 | |||
| 14454 | E | D | D | 5 | |||
| 14455 | E | D | D | 5 | |||
| 5/26/1998 | 14623 | E | D | D | 5 | ||
| 14624 | E | D | D | 5 | |||
| 14625 | E | D | D | 5 | |||
| 14626 | E | D | D | 5 | |||
| 14627 | E | D | D | 5 | |||
| 6/6/1998 | 14628 | E | D | D | 5 | ||
| 14629 | E | D | D | 5 | |||
| 14630 | E | D | D | 5 | |||
| 14631 | E | D | D | 5 | |||
| 14632 | E | D | D | 5 | |||
| 6/15/1998 | 14633 | E | D | D | 5 | ||
| 14634 | E | D | D | 5 | |||
| 14635 | E | D | D | 5 | |||
| 14636 | E | D | D | 5 | |||
| 14637 | E | D | D | 5 | |||
| 11/9/1998 | 15462 | E | D | D | 5 | ||
| 15463 | E | D | D | 5 | |||
| 15464 | E | D | D | 5 | |||
| 15465 | E | D | D | 5 | |||
| 15466 | E | D | D | 5 | |||
| P4 | Giens | 1/8/1998 | 14268 | F | E | E | 6 |
| 14269 | F | E | E | 6 | |||
| 14270 | F | E | E | 6 | |||
| 14271 | F | E | E | 6 | |||
| 14272 | G | F | F | 7 | |||
| 1/21/1998 | 14273 | F | E | E | 6 | ||
| 14274 | F | E | E | 6 | |||
| 14275 | F | E | E | 6 | |||
| 14276 | F | E | E | 6 | |||
| 14277 | F | E | E | 6 | |||
| 2/15/1999 | 15642 | G | F | F | 7 | ||
| 15643 | F | E | E | 6 | |||
| 15644 | F | E | E | 6 | |||
| 15645 | F | E | E | 6 | |||
| 15646 | F | E | E | 6 | |||
| P5 | Giens | 9/22/1998 | 15149 | F | G | G | 8 |
| 15150 | F | G | G | 8 | |||
| 15151 | F | G | G | 8 | |||
| P6 | Giens | 10/26/1998 | 15458 | H | H | H | 9 |
| 15459 | H | H | H | 9 | |||
| 15460 | H | H | H | 9 | |||
| 15461 | H | H | H | 9 | |||
| P7 | Giens | 1/19/1998 | 14354 | I | I | I | 10 |
| 14355 | I | I | I | 10 | |||
| 14356 | I | I | I | 10 | |||
| 14357 | I | I | I | 10 | |||
| 9/22/1998 | 15545 | I | I | I | 10 | ||
| 15546 | I | I | I | 10 | |||
| 15547 | I | I | I | 10 | |||
| 15548 | I | I | I | 10 | |||
| 15549 | I | I | I | 10 | |||
| P8 | Tours | 3/4/1998 | 14462 | J | J | J | 11 |
| 14463 | J | J | J | 11 | |||
| 14464 | F | J | K | 12 | |||
| 14465 | J | J | J | 11 | |||
| 14466 | J | J | J | 11 | |||
| 6/26/1998 | 14754 | K | J | K | 13 | ||
| 14755 | F | J | K | 12 | |||
| 8/11/1998 | 14761 | F | J | K | 12 | ||
| 14762 | F | J | K | 12 | |||
| 14763 | F | J | K | 12 | |||
| 14764 | F | J | K | 12 | |||
| 14765 | F | J | K | 12 | |||
| 9/22/1998 | 15144 | F | J | L | 14 | ||
| 15145 | J | J | J | 11 | |||
| 15146 | F | J | K | 12 | |||
| 15147 | F | J | K | 12 | |||
| 15148 | F | J | K | 12 | |||
| 11/10/1998 | 15551 | F | J | K | 12 | ||
| 15552 | F | J | K | 12 | |||
| 15553 | J | J | J | 11 | |||
| 12/3/1998 | 15554 | J | J | J | 11 | ||
| 15555 | F | J | K | 12 | |||
| 15556 | J | J | J | 11 | |||
| 15557 | F | J | K | 12 | |||
| P9 | Tours | 2/16/1998 | 14358 | L | A | M | 15 |
| 14359 | L | A | M | 15 | |||
| 14360 | L | A | M | 15 | |||
| 14361 | L | A | M | 15 | |||
| 14362 | L | A | M | 15 | |||
| 3/3/1998 | 14467 | M | A | N | 16 | ||
| 14468 | M | A | N | 16 | |||
| 14469 | M | A | N | 16 | |||
| 14470 | M | A | N | 16 | |||
The letters (A, B, and C, etc.) correspond to different electrophoretic patterns generated by each RAPD primer. Note that the electrophoretic patterns were different from one primer to another even if the same letters were used.
IHEM, Institute of Hygiene and Epidemiology, Mycology Section (Brussels, Belgium).
DNA extraction.
The isolates were cultivated to stationary phase in yeast extract-peptone-dextrose broth at 37°C for 2 weeks. After this, the mycelium was collected, washed in sterile distilled water, and ground in liquid nitrogen with a mortar and pestle. Total genomic DNA was extracted as previously described (26), using a DNeasy Plant Mini Prep kit (QIAGEN, Courtaboeuf, France) as recommended by the manufacturer. The DNA concentration was quantified with a TKO 100 fluorimeter (Hoefer Scientific Instruments, San Francisco, Calif.).
PCR amplification.
Three RAPD primers—GC70 (5′-CGGCCACTGT-3′), UBC-701 (5′-CCCAACAACCC-3′), and UBC-703 (5′-CCAACCACCC-3′)—previously selected for their efficiency in discriminating among strains of S. apiospermum (26), were used to amplify total DNA samples. Reaction mixtures (25-μl final volume) consisted of 50 mM KCl, Triton X-100 (0.1%), gelatin (0.2 mg/ml), Tris-HCl buffer (pH 9) containing 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 5 pmol of primer, 25 ng of genomic DNA, and 1 U of Taq DNA polymerase (Appligène-Oncor, Illkirch, France). Amplifications were carried out in a Perkin-Elmer 480 thermocycler programmed for 40 cycles consisting of denaturation (1 min at 94°C), annealing (1 min at 36°C), and elongation (2 min at 72°C). Amplicons were separated by electrophoresis in 1.5% agarose gels in Tris (40 mM)-acetate (10 mM)-EDTA (0.5 mM) buffer (pH 8.0) and visualized by UV transillumination after ethidium bromide staining. Experiments were repeated twice in order to confirm the pattern differences.
Data analysis.
Data were recorded as presence or absence of each amplicon and analyzed using the NT sys-pc program (version 1.80; Exeter Software, Setauket, N.Y.). The similarity degree (SD) was calculated for each couple of genotypes according to the Jacquard coefficient (16). Grouping was carried out by the unweighted pair group method using arithmetic average (UPGMA) cluster analysis (19).
RESULTS
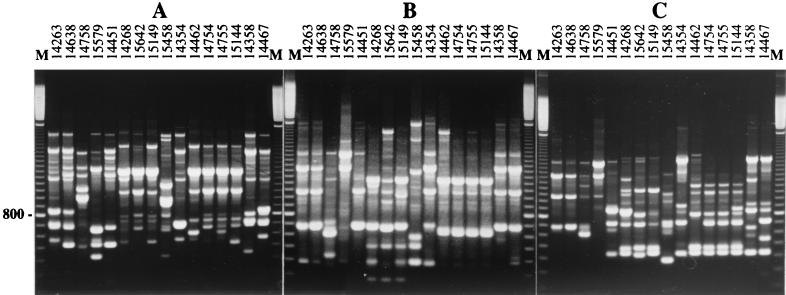
During the sampling period, nine CF patients were found to be colonized by S. apiospermum, and a total of 129 isolates were collected, corresponding to 30 positive sputum samples. Three primers (UBC-701, UBC-703, and GC70), previously selected for their ability to differentiate S. apiospermum isolates (26), were used to amplify the genomic DNA of these isolates. The electrophoretic patterns of resulting amplicons allowed us to identify 16 different genotypes, designated G1 to G16. Each genotype corresponded to a specific combination of electrophoretic patterns obtained with the three selected primers (Fig. 1). Among these primers, UBC-703 was the most discriminating, allowing the differentiation of all but two genotypes. Genotypes G1 and G2, as well as genotypes G12 and G13, could not be differentiated with this primer. These genotypes were differentiated only by using primer GC70 (Fig. 1), whereas genotypes G3, G4, G5, G9, and G10 were distinguished from the others by each of the three primers.
FIG. 1.
Electrophoretic patterns of the 16 genotypes identified in S. apiospermum isolates from CF patients. Amplicons generated by primers GC70 (A), UBC-701 (B), and UBC-703 (C) were separated on 1.5% agarose gels. Lanes M, DNA size markers (100-bp ladder from Amersham Pharmacia-Biotech).
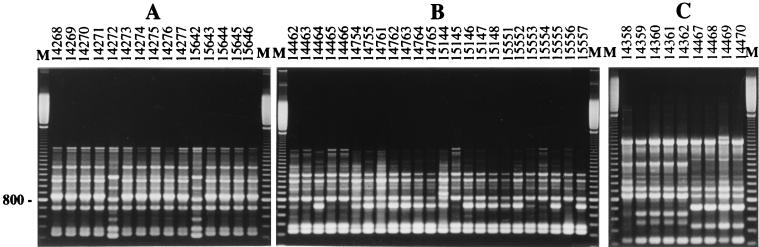
Each genotype was identified in a single patient (Table 2). In addition, a unique genotype was recovered from multiple isolates from two transient carriers, patients P2 and P5, as well as from successive and multiple isolates from two chronically colonized patients (patients P3 and P7). Likewise, although a single sputum sample was available during the sampling period for patient P6, who was colonized by the fungus since 1992, a unique genotype was also recovered from this patient. Two different genotypes were detected for two of the patients, patients P4 and P9, also chronically colonized by the fungus, whereas patients P1 and P8 were infected by three and four genotypes, respectively (Fig. 2). Among the patients colonized by several genotypes, different situations were observed: (i) presence of a quasi-exclusive genotype sporadically accompanied by one or two others, corresponding to only one (patient P1 from Angers) or two (patient P4 from Giens) isolates; (ii) presence of two prevalent genotypes (genotypes G11 and G12) recovered during the full sampling period, with two additional genotypes corresponding each to only one isolate (patient P8 from Tours); or (iii) presence of two distinct genotypes identified successively in the two samples obtained from patient P9, genotype G15 exclusively in the first sample and genotype G16 in the second sample.
FIG. 2.
Electrophoretic patterns of S. apiospermum isolates from CF patients P4 (A), P8 (B), and P9 (C). Amplicons generated by primer UBC-703 were separated on 1.5% agarose gels. Lanes M, DNA size markers (100-bp ladder from Amersham Pharmacia-Biotech).
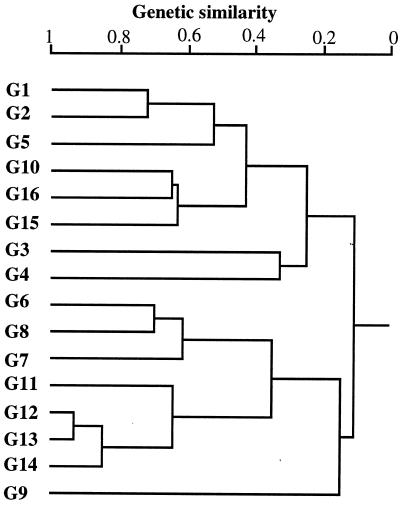
The phylogenetic tree revealed two clusters separated at a normalized distance of 0.11 (Fig. 3). In each cluster, genotypes from different geographic origins were encountered. The different genotypes obtained from a single patient were usually closely related or clustered. For instance, in patient P8, who developed a bronchopulmonary infection, the four genotypes identified (genotypes G11, G12, G13, and G14) were closely related. Likewise, genotypes G15 and G16 from patient P9 and genotypes G6 and G7 from patient P4 were also closely related. Conversely, genotype G3 was far from genotypes G1 and G2 (SD = 0.25), although these three genotypes were recovered from the same patient. The most closely related genotypes were genotypes G12 and G13 (SD = 0.93), and genotype G9, identified in patient P6 from the Giens hospital, was the most distant from the others (SD = 0.15).
FIG. 3.
Dendrogram constructed from RAPD data showing the genetic relationships between the 16 genotypes identified in S. apiospermum isolates from CF patients. The phylogenetic tree was generated from a similarity matrix based on the Jacquard coefficient by using UPGMA cluster analysis. The scale at the top of the figure shows the SD.
DISCUSSION
PCR-based ribotyping was the first molecular method that was proposed for the analysis of Scedosporium isolates (8, 10, 13, 24, 25). However, RAPD, which has been used successfully for numerous epidemiological purposes in medical mycology, also proved its efficiency for the characterization of Scedosporium prolificans isolates (17, 18). In a previous paper, this molecular typing method was compared to multilocus enzyme electrophoresis for the differentiation of strains of S. apiospermum from different origins, including CF patients (26). The best results were obtained by combining three RAPD primers, GC70, UBC-701, and UBC-703, which also presented the best discriminatory power for S. prolificans (17, 18). This three-primer set was therefore used here to investigate the molecular epidemiology of the colonization of the airways by S. apiospermum in nine CF patients monitored in three different hospitals in France (Angers, Giens, and Tours). The mycological survey of these patients resulted in 30 positive sputum samples corresponding to 129 isolates. Sixteen genotypes were identified, with no single genotype common to different patients, allowing us to rule out any transcontamination. In addition, UPGMA analysis did not reveal any clustering according to the geographic origin of the isolates. Interestingly, similar epidemiological features have also been reported for Aspergillus terreus, another filamentous fungus associated with CF (20). Conversely, Williamson et al. (25), using ribotyping of isolates from 29 patients exhibiting pulmonary S. apiospermum infections, showed that patients from the same respiratory medical unit, including CF patients, exhibited one common predominant ribotype, indicating a possible common source of infection. Likewise, the large survey of 255 CF patients infected with B. cepacia carried out by Mahenthiralingam et al. (11) demonstrated the occurrence of widely distributed epidemic genotypes. Although most of the patients were infected with a single strain, seven ribotypes were recovered from multiple patients living in different geographic areas all over the world.
Despite the high polymorphism previously revealed in S. apiospermum by this three-primer set (26), a unique and specific genotype was recovered for five out of the nine patients of our cohort, including two transient carriers. Furthermore, when several genotypes were identified in specimens from the same patient, a predominant genotype was always recovered throughout the sampling period, except for patient P9. For instance, 36 of the 38 isolates obtained from the nine samples collected over 12 months from patient P1 corresponded to the same genotype, genotype G1. Likewise, most of the isolates obtained from the three samples collected over 13 months for patient P4 and from the six samples collected over 9 months for patient P8 belonged to the same genotype.
In addition, the different genotypes identified in specimens from the same patient were usually closely related. For example, genotype G2 was close to the initial genotype, genotype G1, found in patient P1. Likewise, genotypes G11, G12, G13, and G14 from patient P8 were also closely related. The only exception was genotype G3, which was found to be very far from G1 and G2 although it was also recovered from patient P1. Thus, most of the patients studied were colonized by a unique genotype, and when another genotype was detected, it was usually identified in only one clinical sample and corresponded to a unique isolate. A different situation was evidenced in the case of patient P9, for whom two genotypes were successively identified, the first disappearing to the second in a very short interval. However these two genotypes were genetically related (SD = 0.62), indicating the possible derivation of genotype G16 from genotype G15. In agreement with our results, a unique genotype was also identified in each CF patient colonized by A. terreus (20). Likewise, Mahenthiralingam et al. (11) identified only one genotype of B. cepacia from the same patient over 5 years.
Similar methods in our hands led to opposite results for the colonization of the airways of CF patients by A. fumigatus (4). An important polymorphism was revealed from one patient to another, but polymorphism was also observed between different clinical samples from the same patient or even within the same sample, thus confirming previous studies from others (12, 14, 23). This may reflect differences in the occurrence of these fungi in the environment of the patients. A. fumigatus is a widespread ubiquitous mold; all individuals are continuously exposed to the inhalation of airborne conidia belonging to different genotypes, and these conidia find a suitable environment for their development in the airways of CF patients. Conversely, S. apiospermum has been occasionally isolated from soil (5) and has never been recovered from the indoor or outdoor environments of patients (1). In addition, despite the huge biodiversity observed for A. fumigatus, a genotype common to most of the chronically colonized patients tended to settle with the ageing colonization (4), whereas no common genotype was found for patients colonized by S. apiospermum.
In conclusion, these results confirmed our previous study, which did not reveal any clustering according to the origin of the isolates. It has to be specified, however, that if RAPD is well suited for an accurate differentiation of the strains, this method is perhaps too discriminating to reveal clustering. In addition, our results demonstrated the clonality of the S. apiospermum organisms colonizing the airways. Unlike A. fumigatus, and despite the antifungal treatments, a unique genotype was almost recovered from our patients, probably in relation to the low occurrence of the fungus in the environments of the patients and its poor sensitivity to antifungal drugs (7). This study therefore reinforces the need for the development of new antifungal drugs that are more efficient against this filamentous fungus.
Acknowledgments
This work was supported by a grant from the Programme Hospitalier de Recherche Clinique du Ministère des Affaires Sociales, de la Santé et de la Ville (PHRC 1997).
REFERENCES
- 1.Beguin, H., and N. Nolard. 1994. Mould biodiversity in homes. Air and surface analysis of 130 dwellings. Aerobiologia 10:157-166. [Google Scholar]
- 2.Cimon, B., J. Carrère, J. P. Chazalette, J. L. Giniès, P. Six, J. F. Vinatier, D. Chabasse, and J. P. Bouchara. 1995. Fungal colonization and immune response to fungi in cystic fibrosis. J. Mycol. Med. 5:211-216. [Google Scholar]
- 3.Cimon, B., J. Carrère, J. F. Vinatier, J. P. Chazalette, D. Chabasse, and J. P. Bouchara. 2000. Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 19:53-56. [DOI] [PubMed] [Google Scholar]
- 4.Cimon, B., F. Symoens, R. Zouhair, D. Chabasse, N. Nolard, A. Defontaine, and J. P. Bouchara. 2001. Molecular epidemiology of airway colonisation by Aspergillus fumigatus in cystic fibrosis patients. J. Med. Microbiol. 50:367-374. [DOI] [PubMed] [Google Scholar]
- 5.Cremer, G., and P. Boiron. 1996. Epidemiology and biology of Scedosporium species. J. Mycol. Med. 6:165-171. [Google Scholar]
- 6.Dupont, B., L. Improvisi, and O. Ronin. 1991. Aspects épidémiologiques et cliniques des infections à Scedosporium et Pseudallescheria. J. Mycol. Med. 1:33-42. [Google Scholar]
- 7.Hennequin, C., N. Benailly, C. Silly, M. Sorin, P. Scheinmann, G. Lenoir, J. L. Gaillard, and P. Berche. 1997. In vitro susceptibilities to amphotericin B, itraconazole, and miconazole of filamentous fungi isolated from patients with cystic fibrosis. Antimicrob. Agents Chemother. 41:2064-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issakainen, J., J. Jalava, E. Eerola, and C. K. Campbell. 1997. Relatedness of Pseudallescheria, Scedosporium and Graphium pro parte based on SSU rDNA sequences. J. Med. Vet. Mycol. 35:389-398. [PubMed] [Google Scholar]
- 9.Koch, C., and N. Hoiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 10.Lennon, P. A., C. R. Cooper, L. F. L. Salkins, and S. B. Lee. 1994. Ribosomal DNA internal transcribed spacer analysis supports synonymy of Scedosporium inflatum and Lomentospora prolificans. J. Clin. Microbiol. 32:2413-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuvéglise, C., J. Sarfati, J. P. Debeaupuis, H. Vu Thien, J. Just, G. Tournier, and J. P. Latgé. 1997. Longitudinal study of Aspergillus fumigatus strains isolated from cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:747-750. [DOI] [PubMed] [Google Scholar]
- 13.Rainer, J., G. S. de Hoog, M. Wedde, Y. Gräser, and S. Gilges. 2000. Molecular variability of Pseudallescheria boydii. J. Clin. Microbiol. 38:3267-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rath, P. M., F. Ratjen, and R. Ansorg. 1997. Genetic diversity among isolates of Aspergillus fumigatus in patients with cystic fibrosis. Zentbl. Bakteriol. 285:450-455. [DOI] [PubMed] [Google Scholar]
- 15.Rippon, J. W. 1988. Pseudallescheriasis, p. 651-680. In J. W. Rippon (ed.), Medical mycology. The pathogenic fungi and the pathogenic Actinomycetes, 3rd ed. W. B. Saunders, Philadelphia, Pa.
- 16.Romesburg, H. C. 1984. Cluster analysis for researchers. Lifetime Learning Publications, Belmont, Calif.
- 17.Ruis-Diez, B., F. Martin-Diez, J. L. Rodriguez-Tudela, M. Alvarez, and J. V. Martinez-Suarez. 1997. Use of random amplification of polymorphic DNA (RAPD) and PCR-fingerprinting for genotyping a Scedosporium prolificans (inflatum) outbreak in four leukemic patients. Curr. Microbiol. 35:186-190. [DOI] [PubMed] [Google Scholar]
- 18.San Millan, R., G. Quindos, J. Garaizar, R. Salesa, J. Guarro, and J. Ponton. 1997. Characterization of Scedosporium prolificans clinical isolates by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 35:2270-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sneath, P. H., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, Calif.
- 20.Symoens, F., J. P. Bouchara, S. Heinemann, and N. Nolard. 2000. Molecular typing of Aspergillus terreus isolates by random amplification of polymorphic DNA. J. Hosp. Infect. 44:273-280. [DOI] [PubMed] [Google Scholar]
- 21.Tadros, T. S., K. A. Workowski, R. J. Siegel, S. Hunter, and D. A. Schwartz. 1998. Pathology of hyalohyphomycosis caused by Scedosporium apiospermum (Pseudallescheria boydii): an emerging mycosis. Hum. Pathol. 29:1266-1272. [DOI] [PubMed] [Google Scholar]
- 22.Travis, L. B., G. D. Roberts, and W. R. Wilson. 1985. Clinical significance of Pseudallescheria boydii: a review of 10 years' experience. Mayo Clin. Proc. 60:531-537. [DOI] [PubMed] [Google Scholar]
- 23.Verweij, P. E., J. F. G. M. Meis, J. Sarfati, J. A. A. Hoogkamp-Korstanje, J.-P. Latgé, and W. J. G. Melchers. 1996. Genotypic characterization of sequential Aspergillus fumigatus isolates from patients with cystic fibrosis. J. Clin. Microbiol. 34:2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wedde, M., D. Müller, K. Tintelnot, G. S. de Hoog, and U. Stahl. 1998. PCR-based identification of clinically relevant Pseudallescheria/Scedosporium strains. Med. Mycol. 36:61-67. [PubMed] [Google Scholar]
- 25.Williamson, E. C. M., D. Speers, I. H. Arthur, G. Harnett, G. Ryan, and T. J. J. Inglis. 2001. Molecular epidemiology of Scedosporium apiospermum infection determined by PCR amplification of ribosomal intergenic spacer sequences in patients with chronic lung disease. J. Clin. Microbiol. 39:47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zouhair, R., A. Defontaine, C. Ollivier, B. Cimon, F. Symoens, J. N. Hallet, J. Deunff, and J. P. Bouchara. 2001. Typing of Scedosporium apiospermum strains by multilocus enzyme electrophoresis and random amplification of polymorphic DNA. J. Med. Microbiol. 50:925-932. [DOI] [PubMed] [Google Scholar]