Abstract
To improve the diagnosis of invasive aspergillosis (IA), we developed a LightCycler PCR assay targeted to Aspergillus fumigatus and A. flavus mitochondrial DNA. To avoid contamination, fully automated nucleic acid extraction with the MagNA Pure LC apparatus was used. The linearity of the results was achieved over a 6-log range of input A. fumigatus DNA, from 0.3 ng to 3 fg/10 μl of water. We retrospectively compared the LightCycler PCR and an enzyme-linked immunosorbent assay for the detection of galactomannan (GM) in serum from 14 patients with IA. The GM assay was more frequently positive (57 of 109; 52%) than the PCR assay (49 of 109; 45%). The LightCycler PCR assay, combined with automated DNA extraction, could be used in association with the GM assay to improve the reliability of IA diagnosis.
The incidence of invasive aspergillosis (IA) is difficult to assess, as at least 30% of the cases that occur remain undiagnosed and untreated at death (7). Two approaches have been investigated to improve the sensitivity of aspergillosis detection by noninvasive techniques; these are antigen detection and DNA detection (13). Among the antigen tests, detection of galactomannan (GM) by an enzyme-linked immunosorbent assay (ELISA) has been shown to have good sensitivity, but a false-positivity rate of 10 to 15% has been reported (5, 16). Nevertheless, this method is now included in the IA diagnosis criteria (1).
PCR technology is not as acceptable a diagnostic tool as the GM assay because very divergent results have been reported (3, 9). These discrepancies are most likely due to technical reasons because the PCR techniques used are not standardized. Real-time PCR assays dramatically decrease the risk of false-positive results. Indeed, contamination with previously amplified products is reduced because the reaction tubes need not be opened following amplification, thus avoiding potential contamination of the environment with amplicons (6, 15). We therefore developed a PCR assay based on the LightCycler technology (19). We also employed a fully automated nucleic acid extraction technique with the MagNA Pure LC apparatus (12). This apparatus can purify DNA from a broad variety of samples by magnetic bead technology, thus eliminating the need for vacuum pumps, centrifugation, or other manual steps that may result in cross contamination. We subsequently compared the performance of the GM assay and the real-time PCR test with samples from selected patients.
LightCycler PCR test development.
The LightCycler PCR test was targeted at the Aspergillus fumigatus mitochondrial gene (GenBank accession number L37095) (4). The amplicon comprises a 91-bp fragment. The two hybridization probes (TibMolBiol, Berlin, Germany) used are the 24-mer 5′-CTGTTAGTGCGGGAGTTCAAAXTCT-3′, where X represents the internal labeling of the T with LC-Red 640, and the 28-mer 5′-CTGAGCTAATTTCTTTCAACCCAAGGGA-3′, which is labeled at the 3′ end with fluorescein isothiocyanate. Because of the thermodynamic constraints on the choice of the primers and probes, the LC-Red 640 hybridization probe also serves as the sense primer for amplification and the 22-mer antisense primer sequence is 5′-AACACCTGACCTTTCGCGTGTA-3′(MWG, Courtaboeuf, France). For quantification, a single 10-fold serial dilution of A. fumigatus DNA (strain IP 2279; Institut Pasteur Collection, Paris, France) was made, ranging from 0.3 ng to 3 fg/10 μl. The last dilution of A. fumigatus DNA that was consistently positive was included in each amplification run as a positive control.
LightCycler PCRs were set up in a final volume of 20 μl with the Fast DNA Master Hybridization Probes Kit (Roche Molecular Biochemicals, Meylan, France), including heat-activable Taq polymerase, with 4.5 mM MgCl2, 0.25 U of UDG (Biolabs, St. Quentin, France), each primer at 0.5 mM, the fluorescein isothiocyanate-labeled probe at 0.25 mM, and 10 μl of extracted DNA sample. After one step at 50°C to allow the action of UDG and another one at 95°C for 8 min to activate the Taq polymerase and inactivate the UDG, the samples were cycled 50 times (denaturation at 95°C for 5 s with a ramp rate of 20°C/s, annealing at 60°C for 10 s with a ramp rate of 20°C/s, and extension at 72°C for 15 s with a ramp rate of 2°C/s). The amplification was done with a LightCycler Instrument (Roche Molecular Biochemicals). Fluorescence was measured once every cycle immediately after the 60°C incubation (extension step). Fluorescence curves were analyzed with the LightCycler software, version 3.5. Quantitative results were expressed by determination of the threshold of detection, or crossing point (Cp), which marked the cycle at which the fluorescence of the sample became significantly different from the baseline signal. The Cp is expressed as a fractional cycle number and is defined as the maximum of the second derivative from the fluorescence curve of the sample. Automated calculation is done by the second derivative maximum method. Cycles 15 to 50 were selected for calculation of the Cp. Each sample was tested in duplicate. The final Cp was the mean of the two results.
Sample processing.
Patients at risk for IA were screened once or twice weekly for the presence of GM. Upon the receipt of a patient's serum, the ELISA was performed on a part of the serum as previously described (3) and 1 ml of serum was collected in a screw-cap tube and stored at −80°C. According to the manufacturer's instructions, a GM assay result is positive when the GM index (the sample's optical density divided by the optical density of 1 ng of GM per ml) is ≥1.5 and doubtful between 1 and 1.5.
DNA extraction of serum was performed with the MagNA Pure LC apparatus (Roche Biochemicals). This benchtop instrument can extract 32 samples in parallel. The total nucleic acid isolation kit (Roche Biochemicals) was used. We used a starting volume of 500 μl, eluted in 50 μl, and 10 μl was used for the PCR. After the DNA extraction was completed, eluted DNA and PCR reagents were automatically dispensed into PCR capillaries with an integrated PCR setup procedure. Operator manipulation was thus restricted to introduction of the reagents and samples into the apparatus. All of the mechanical methods used to prevent DNA contamination as previously described (4) were also included.
Blood was collected from three patients in plain and EDTA-containing tubes on the same day to compare efficiencies of Aspergillus DNA recovery. One, five, and eight pairs of specimens from patients 8, 13, and 14, respectively (Table 1), were obtained on consecutive days. Plasma and white cells pellets were obtained from the EDTA-treated blood specimens as previously described (6). White cell pellets were resuspended in 500 μl of water, and the DNA was extracted from three different fractions, i.e., serum, plasma, and white cell pellets, as described above.
TABLE 1.
Comparative results of GM antigen ELISA and LightCycler PCR test obtained with serum samples from 20 patients with IA
| Patient no. | Sex, age (yr) | Mycology | EORTC criteriaa | No. of samples
|
||
|---|---|---|---|---|---|---|
| Total (n = 158) | GM positive (n = 71) | PCR positive (n = 49) | ||||
| 1 | F, 57 | A. fumigatus | P | 6 | 2 | 2 |
| 2 | M, 51 | A. fumigatus | P | 5 | 3 | 5 |
| 3 | M, 56 | A. fumigatus | P | 2 | 2 | 1 |
| 4 | M, 41 | Positive microscopy | P | 9 | 8 | 2 |
| 5 | F, 44 | A. fumigatus | C | 11 | 4 | 1 |
| 6 | F, 32 | A. fumigatus | C | 14 | 1 | 2 |
| 7 | M, 72 | Positive microscopy | P | 10 | 5 | 3 |
| 8 | F, 34 | A. fumigatus | C | 19 | 7 | 12 |
| 9 | F, 46 | A. flavus | C | 3 | 1 | 1 |
| 10 | M, 78 | Negative | P | 4 | 4 | 2 |
| 11 | M, 21 | Negative | P | 5 | 4 | 4 |
| 12 | F, 43 | Negative | P | 5 | 1 | 2 |
| 13 | F, 68 | Negative | P | 3 | 3 | 3 |
| 14 | M, 47 | Negative | P | 13 | 11 | 9 |
| 15 | M, 49 | A. fumigatus | P | 3 | 1 | 0 |
| 16 | F, 55 | Negative | P | 6 | 2 | 0 |
| 17 | M, 43 | Positive microscopy | P | 8 | 4 | 0 |
| 18 | M, 19 | Negative | P | 8 | 4 | 0 |
| 19 | F, 62 | Negative | P | 9 | 2 | 0 |
| 20 | M, 40 | Negative | P | 15 | 2 | 0 |
EORTC, European Organization for Research and Treatment of Cancer. C, proven; P, probable.
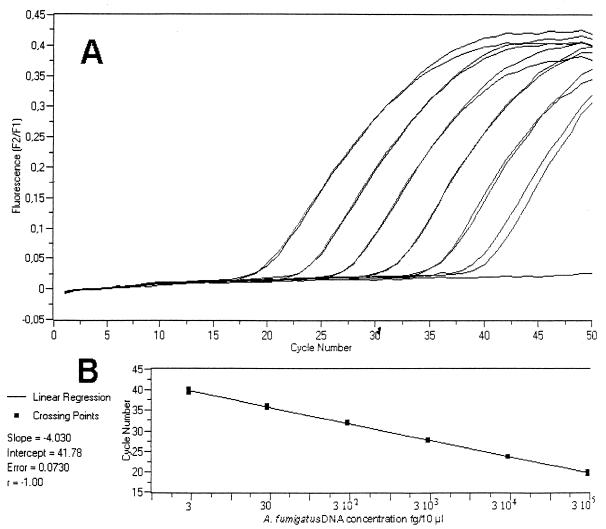
Figure 1 shows the amplification of 10-fold serial dilutions of purified A. fumigatus DNA with fluorescence plotted against cycle number. Linearity was achieved over a 6-log range of input DNA amounts (from 0.3 ng to 3 fg/10 μl), showing the wide dynamic range and accuracy of the PCR assay. Reproducibility was determined by testing 10-fold dilutions of A. fumigatus DNA in five independent runs. The coefficient of variation was ≤1.7%. Similar results were observed with A. flavus DNA (data not shown).
FIG. 1.
Real-time quantitative LightCycler PCR test with fluorescent energy transfer. (A) Duplicate amplification plots obtained for A. fumigatus DNA dilutions of 3 × 105 (left) to 3 (right) fg/10 μl. Each slope corresponds to a given input target quantity. (B) Plot of the duplicate Cp against the input target quantity (common log scale) showing the linearity of the results from 3 × 105 (right) to 3 (left) fg. The computer-calculated correlation coefficient is 1.
Two hundred ten serum samples were tested with the GM ELISA and the real-time quantitative LightCycler PCR. Ten patients, all having undergone allogeneic bone marrow transplantation without any clinical history of aspergillosis, were both GM and PCR negative (42 samples). Additionally, 20 serum samples from healthy individuals were all PCR negative.
We retrospectively studied 20 patients mainly with hematological malignancies classified, in accordance with the European Organization for Research and Treatment of Cancer criteria (1), as having confirmed (n = 4) or probable (n = 16) IA (Table 1). These 20 patients had a GM index of >1.5 at least once. Fourteen of them were PCR positive at least once. For the patients who were positive by both tests, the GM test was more frequently positive (57 of 109; 52%) than was the LightCycler PCR test (49 of 109; 45%). In contrast to the GM titers, we did not observe any frank increase in the DNA load during the course of the disease. Most of the Cps were between 36 and 40 cycles. Therefore, even before death, the DNA concentration was never greater than 30 fg/ml of serum. The different blood fractions (plasma, serum, and white cell pellet) yielded similar DNA quantitations (data not shown).
A crucial point in the amplification of fungal DNA for diagnosis is the possibility of reagent contamination with fungal DNA (10, 14, 17). To anticipate and overcome this problem, all reagents were irradiated for 8 h with UV light in the MagNa Pure LC apparatus during the decontamination step. Therefore, we can exclude the possibility of false-positive results in this study.
Comparisons of antigen and DNA detection tests have already been reported (2, 3, 8, 11). Some investigators (8) have compared nested PCR and GM detection with the latex test (Pastorex; Bio-Rad), a relatively insensitive test (13). Other investigators have demonstrated the superiority of a nested PCR test over the GM ELISA, but most of the patients tested had aspergilloma and not IA (11). The present results are similar to those we previously reported for a competitive PCR assay for human disease (3) and in an experimental animal model of IA (2). The fact that antigen detection tests can be more sensitive than PCR is not surprising if the fungus releases large amounts of antigen that can be easily detected compared to the small quantity of circulating DNA that is harder to detect even after amplification. Indeed, the DNA load, as estimated by quantitative PCR, was always low, i.e., less than 30 fg/ml of serum. There is a suggestion that the host response to the fungus may prevent release of fungal DNA from the lesion to the bloodstream (18). However, the origin and kinetics of fungal DNA release and its circulation require additional study.
Our results confirm that serum is an appropriate source for A. fumigatus DNA amplification and should be preferred to white cells (6). If phagocytosed conidia or hyphae were responsible for the positive PCR results, blood cultures would be positive, which is rarely observed during IA (7). Moreover, the number of white cells from patients with IA is often a limiting factor because most of them are neutropenic (7). Therefore, serum should be assayed instead of white cell pellets, whose preparation is cumbersome. Serum can also be used for other purposes, e.g., antigenemia detection, and therefore decreases the need to draw blood for routine diagnosis and for epidemiological studies.
The real-time quantitative LightCycler PCR assay used in this study, combined with automated DNA extraction from serum, provides better reliability and safety than the competitive PCR test previously used. We think that the real-time PCR test can be used to improve the confidence of IA diagnosis, especially when a false-positive GM result might lead to unnecessary antifungal therapy. Determination of whether the real-time PCR test can be more useful than other diagnostic tools for the early diagnosis of IA requires prospective studies.
Acknowledgments
The data in this study were generated with a MagNA Pure LC apparatus kindly provided by Roche Molecular Biochemicals, Meylan, France.
We thank Richard Calderone, Georgetown University, for critical reading of the manuscript.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect Dis 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. J., S. de Marie, D. Willemse, H. A. Verbrugh, and I. A. Bakker-Woudenberg. 2000. Quantitative galactomannan detection is superior to PCR in diagnosing and monitoring invasive pulmonary aspergillosis in an experimental rat model. J. Clin. Microbiol. 38:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretagne, S., J.-M. Costa, E. Bart-Delabesse, N. Dhédin, C. Rieux, and C. Cordonnier. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction in diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 4.Bretagne, S., J. M. Costa, A. Marmorat-Khuong, F. Poron, C. Cordonnier, M. Vidaud, and J. Fleury-Feith. 1995. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J. Clin. Microbiol. 33:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretagne, S., A. Marmorat-Khuong, M. Kuentz, J. P. Latgé, E. Bart-Delabesse, and C. Cordonnier. 1997. Serum Aspergillus galactomannan antigen testing by sandwich ELISA: practical use in neutropenic patients. J. Infect. 35:7-15. [DOI] [PubMed] [Google Scholar]
- 6.Costa, C., D. Vidaud, M. Olivi, E. Bart-Delabesse, M. Vidaud, and S. Bretagne. 2001. Development of two real-time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. J. Microbiol. Methods 44:263-269. [DOI] [PubMed] [Google Scholar]
- 7.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto, A., Y. Yamakami, P. Kamberi, E. Yamagata, R. Karashima, H. Nagaoka, and M. Nasu. 1998. Comparison of PCR, (1→3)-β-d-glucan and galactomannan assays in sera of rats with experimental invasive aspergillosis. J. Clin. Lab. Anal. 12:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebart, H., J. Loffler, C. Meisner, F. Serey, D. Schmidt, A. Bohme, H. Martin, A. Engel, D. Bunje, W. V. Kern, U. Schumacher, L. Kanz, and H. Einsele. 2000. Early detection of aspergillus infection after allogeneic stem cell transplantation by polymerase chain reaction screening. J. Infect. Dis 181:1713-1719. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez, L., S. Smalls, and R. Ignar. 2000. Use of PCR analysis for detecting low levels of bacteria and mold contamination in pharmaceutical samples. J. Microbiol. Methods 41:259-265. [DOI] [PubMed] [Google Scholar]
- 11.Kawamura, S., S. Maesaki, T. Noda, Y. Hirakata, K. Tomono, T. Tashiro, and S. Kohno. 1999. Comparison between PCR and detection of antigen in sera for diagnosis of pulmonary aspergillosis. J. Clin. Microbiol. 37:218-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler, H. H., G. Muhlbauer, E. Stelzl, E. Daghofer, B. I. Santner, and E. Marth. 2001. Fully automated nucleic acid extraction: MagNA Pure LC. Clin. Chem. 47:1124-1136. [PubMed] [Google Scholar]
- 13.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeffler, J., H. Hebart, R. Bialek, L. Hagmeyer, D. Schmidt, F. P. Serey, M. Hartmann, J. Eucker, and H. Einsele. 1999. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 37:1200-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertens, J., J. Verhaegen, K. Lagrou, J. Van Eldere, and M. Boogaerts. 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood 97:1604-1610. [DOI] [PubMed] [Google Scholar]
- 17.Rimek, D., A. P. Garg, W. H. Haas, and R. Kappe. 1999. Identification of contaminating fungal DNA sequences in Zymolyase. J. Clin. Microbiol. 37:830-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verweij, P. E., C. M. Weemaes, J. H. Curfs, S. Bretagne, and J. F. Meis. 2000. Failure to detect circulating Aspergillus markers in a patient with chronic granulomatous disease and invasive aspergillosis. J. Clin. Microbiol. 38:3900-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]