Klaassen et al. (5) recently compared amplified fragment length polymorphism (AFLP) with pulsed-field gel electrophoresis (PFGE) as DNA fingerprinting methods for Clostridium difficile. It is known that certain C. difficile strains are problematic to fingerprint by PFGE due to DNA degradation (2, 6). Corkill et al. suggested that such strains could be typed by PFGE if 50 μM thiourea was added to the electrophoresis buffer (3). However, Klaassen et al. reported that although DNA degradation was reduced by this new method, analysis of some C. difficile strains remained difficult due to residual DNA breakdown.
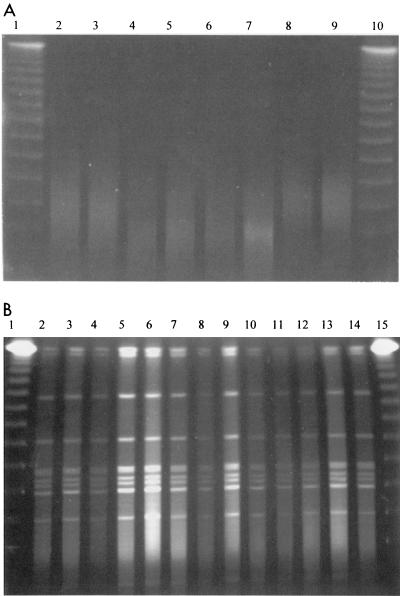
We agree with the conclusion by Klaassen et al. that strains from hospital A represented clonal expansion of a single degradation-susceptible strain, but we were surprised at their assertion that previous data do not exist to support widespread dissemination of C. difficile isolates. In England and Wales, the Public Health Laboratory Service Anaerobic Reference Unit reported on the most common PCR ribotypes and established that ribotype 1 was present in 33 of 58 hospitals and accounted for 55% of infections in the United Kingdom (1, 7). A recent epidemiological investigation within our own hospital established the presence of this same United Kingdom epidemic strain. Ribotype 1 represented approximately 80% of all C. difficile isolates (4) and was found to be degradation susceptible during PFGE analysis. We have now modified our PFGE method to allow analysis of formerly degradation-susceptible C. difficile strains (Fig. 1A and B). Our optimized method differs significantly from that used by Klaassen et al. in several key areas, and we suggest that similar modifications might improve the typeability of their strains.
FIG. 1.
PFGE DNA fingerprints of 8 representative ribotype 1 C. difficile isolates (lanes 2 to 9) obtained by using an unmodified PFGE protocol (Fig. 1A) and 13 representative ribotype 1 isolates (lanes 2 to 14) obtained by using our modified PFGE protocol (Fig. 1B). The markers present in lanes 1 and 10 (Fig. 1A) and lanes 1 and 15 (Fig. 1B) are bacteriophage lambda concatemers. Gel images were generated by an ImageMaster VDS camera (Pharmacia) and were sized by using Adobe Photoshop software.
C. difficile strains were grown overnight in Schaedler's anaerobic broth as opposed to on agar plates to discourage mature spore formation which contributes to low yields of poor-quality DNA. In addition, more bacteria were incorporated into the agarose plugs (approximately 2.0 McFarland standards) to ensure that sufficient DNA remained present in the event of residual DNA degradation. Lysozyme was incorporated into the agarose plugs (10 μg/ml) and incubated at 37°C for 1 h to ensure optimal bacterial lysis. The concentration of proteinase K was increased to 5 mg/ml to allow for the increased bacterial load. It was found that adequate bacterial digestion was achieved if blocks were exposed to proteinase K for a minimum of 16 h at 55°C with fresh solutions added after approximately 8 h. We found that 200 μM thiourea must be present in both the agarose gel and the electrophoresis buffer to ensure minimal DNA degradation.
We have found that the random amplified polymorphic DNA technique and PFGE have similar discriminatory powers (unpublished results) and therefore consider PFGE to be a valuable tool in epidemiological analysis of C. difficile strains. We hope that our PFGE protocol modifications will allow a full evaluation of PFGE, AFLP, and other C. difficile molecular fingerprinting techniques.
REFERENCES
- 1.Brazier, J. S. 1998. The epidemiology and typing of Clostridium difficile. J. Antimicrob. Chemother. 41(Suppl.):C47-C57. [DOI] [PubMed] [Google Scholar]
- 2.Chachaty, E., P. Saulnier, A. Martin, N. Mario, and A. Andremont. 1994. Comparison of ribotyping, pulsed-field gel electrophoresis and random amplified polymorphic DNA for typing Clostridium difficile strains. FEMS Microbiol. Lett. 122:61-68. [DOI] [PubMed] [Google Scholar]
- 3.Corkill, J. E., R. Graham, C. A. Hart, and S. Stubbs. 2000. Pulsed-field gel electrophoresis of degradation-sensitive DNAs from Clostridium difficile PCR ribotype 1 strains. J. Clin. Microbiol. 38:2791-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fawley, W. N., and M. H. Wilcox. 2001. Molecular epidemiology of endemic Clostridium difficile infection. Epidemiol. Infect. 126:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klaassen, C. H., H. A. van Haren, and A. M. Horrevorts. 2002. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J. Clin. Microbiol. 40:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristjansson, M., M. H. Samore, D. N. Gerding, P. C. DeGirolami, K. M. Bettin, A. W. Karchmer, and R. D. Arbeit. 1994. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J. Clin. Microbiol. 32:1963-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]