Abstract
The sixth pandemic of cholera and, presumably, the earlier pandemics were caused by the classical biotype of Vibrio cholerae O1, which was progressively replaced by the El Tor biotype representing the seventh cholera pandemic. Although the classical biotype of V. cholerae O1 is extinct, even in southern Bangladesh, the last of the niches where this biotype prevailed, we have identified new varieties of V. cholerae O1, of the El Tor biotype with attributes of the classical biotype, from hospitalized patients with acute diarrhea in Bangladesh. Twenty-four strains of V. cholerae O1 isolated between 1991 and 1994 from hospitalized patients with acute diarrhea in Matlab, a rural area of Bangladesh, were examined for the phenotypic and genotypic traits that distinguish the two biotypes of V. cholerae O1. Standard reference strains of V. cholerae O1 belonging to the classical and El Tor biotypes were used as controls in all of the tests. The phenotypic traits commonly used to distinguish between the El Tor and classical biotypes, including polymyxin B sensitivity, chicken cell agglutination, type of tcpA and rstR genes, and restriction patterns of conserved rRNA genes (ribotypes), differentiated the 24 strains of toxigenic V. cholerae O1 into three types designated the Matlab types. Although all of the strains belonged to ribotypes that have been previously found among El Tor vibrios, type I strains had more traits of the classical biotype while type II and III strains appeared to be more like the El Tor biotype but had some classical biotype properties. These results suggest that, although the classical and El Tor biotypes have different lineages, there are possible naturally occurring genetic hybrids between the classical and El Tor biotypes that can cause cholera and thus provide new insight into the epidemiology of cholera in Bangladesh. Furthermore, the existence of such novel strains may have implications for the development of a cholera vaccine.
New epidemic strains of toxigenic Vibrio cholerae have appeared at least twice in recent human history (10). Strains of the classical biotype, which had probably been responsible for most of the epidemic disease in the 19th century and much of the 20th century, were largely replaced as the predominant cause of epidemic cholera by strains of the El Tor biotype in most of the regions where cholera is endemic, beginning in 1961. However, the classical biotype strains reemerged as a predominant epidemic strain in parts of Bangladesh in 1982 (8, 25) and coexisted with the El Tor strains, causing disease until 1993. A second new epidemic strain, carrying the O139 rather than the O1 antigen, emerged in southern Asia in 1992 (7, 24). The O139 and El Tor O1 strains continue to cause epidemics of cholera, and there are indications that the incidence of cholera due to the O139 serogroup is on the rise in parts of India and Bangladesh.
The classical and El Tor biotypes of V. cholerae are closely related in their O-antigen biosynthetic genes (21, 31), although these two biotypes differ in many other regions of their genomes (2, 16, 17, 29, 30). Thus, O1 El Tor strains might have arisen following transfer of O1 antigen biosynthetic genes into a previously unknown environmental strain. Conversely, O139 and O1 El Tor strains are closely related in most parts of their genomes but carry different O-antigen genes, suggesting the transfer of O139-specific genes from an unknown donor into a recipient El Tor strain (3, 28). Similar conclusions about gene transfer have emerged from comparisons of serogroups and sequences of diagnostic housekeeping genes of nonepidemic isolates (2).
In this study, we have identified a new variety of V. cholerae O1 that appears to be a hybrid of the classical and El Tor biotypes from hospitalized patients with acute diarrhea. The phenotypic traits that distinguish the classical and El Tor biotypes of V. cholerae O1 and important discriminating genotypic characteristics of these strains are reported here, and the implications of the existence of such novel strains, especially in relation to cholera vaccine development, are described.
MATERIALS AND METHODS
Twenty-four strains of V. cholerae isolated between 1991 and 1994 from hospitalized patients with acute diarrhea in the Matlab hospital, 45 km south of Dhaka, Bangladesh, were included in this study. The basis of a retrospective reexamination of these strains was their unusual response to polymyxin B (50 U), chicken cell agglutination (CCA), Voges-Proskauer (VP) reaction, and sensitivity to group IV and V phages, all of which are phenotypic traits commonly used to differentiate between the classical and El Tor biotypes. The 24 strains were reexamined for the above phenotypic characteristics by standard procedures.
The presence of the ctxA gene and the variants of the classical and El Tor tcpA genes was determined by a multiplex PCR assay (18). The expected size of the PCR amplicons was ascertained by electrophoresis in agarose gels. The identities of all PCR products were further verified with specific oligonucleotide probes. The probes for El Tor and classical biotype-specific CTX prophage repressor rstR were SacI-XbaI fragments of pHK1 and pHK2, respectively (19). The acfB gene probe was prepared from the PCR amplicon with previously reported acfB-specific primers (13). The rRNA gene probe consisted of a 7.5-kb BamHI fragment of Escherichia coli rRNA clone pKK3535 (5). Colony blots or Southern blots were prepared with nylon filters (Hybond; Amersham International plc., Aylesbury, United Kingdom) by standard methods (27). The probes were labeled by random priming (14) with a random-primer DNA labeling kit (Bethesda Research Laboratories, Gaithersburg, Md.) and [α-32P]dCTP (3,000 Ci/mmol; Amersham). Colony blots and Southern blots were hybridized with the probes and autoradiographed as described previously (11-13).
RESULTS
The commonly used phenotypic traits used to distinguish between the El Tor and classical biotypes of V. cholerae differentiated the 24 strains into three types (Table 1), which we classified as Matlab types I, II, and III (named after the place where these strains were first isolated). Matlab type I included two strains belonging to the Inaba serotype that were resistant to both the El Tor-specific group IV and the classical biotype-specific group V phages, negative by the CCA and VP tests (both are classical traits), and resistant to polymyxin B (an El Tor trait). Matlab type II included one strain belonging to the Ogawa serotype that was sensitive to the group IV phage but showed negative responses in the CCA and VP tests and was sensitive to polymyxin B, all of which are classical biotype characteristics. Matlab type III included 21 Ogawa strains that showed the sensitivity to phages and polymyxin B typical of the El Tor biotype but were negative by the CCA and VP tests (both classical biotype traits).
TABLE 1.
Phenotypic traits of Matlab types I, II, and III of toxigenic V. cholerae O1 isolated from patients hospitalized with acute secretory diarrhea in Bangladesh
| Type | No. of strains | No. of strains of serotype:
|
VP testa | Sensitivity to poly- myxin B (50 U)b | CAAa | Phage sensitivitya
|
||
|---|---|---|---|---|---|---|---|---|
| Inaba | Ogawa | Group IV (El Tor biotype specific) | Group V (classical biotype specific) | |||||
| Matlab I | 2 | 2 | 0 | − | R | − | R | R |
| Matlab II | 1 | 0 | 1 | − | S | − | S | R |
| Matlab III | 21 | 0 | 21 | − | R | − | S | R |
| El Tor MAK757 | 1 | 0 | 1 | + | R | + | S | R |
| Classical 154 | 1 | 0 | 1 | − | S | − | R | S |
+, positive; −, negative.
Abbreviations: R, resistant; S, sensitive.
Genotypically, all of the strains carried the ctxA gene, a constituent gene of the CTX prophage that encodes cholera toxin, and acfB and tcpA, which are located in different gene clusters (acf and tcp gene clusters) on the V. cholerae pathogenicity island. The type I strains appeared to belong more to the classical biotype because they carried the tcpA gene and the CTX prophage repressor gene (rstR) of the classical type (Table 2). The tcpA gene of the single type II strain was of the classical type, while the rstR gene was of the El Tor type. The six representative strains of V. cholerae representing Matlab III also carried the tcpA gene of the classical type. Five of the strains had the El Tor-type rstR gene, while one carried both the El Tor and classical rstR types.
TABLE 2.
Genotypic traits of V. cholerae O1 strains isolated from hospitalized patients with acute diarrhea in Bangladesha
| Strain | Matlab type | Yr of isolation | tcpA PCR | ctxA PCR | acfB (probe) | rstR (probe) |
|---|---|---|---|---|---|---|
| MJ-1236 | I | 1994 | C | + | + | C |
| MJ-1485 | I | 1994 | C | + | + | C |
| MG-116226 | II | 1991 | C | + | + | E |
| MG-116025 | III | 1991 | C | + | + | E |
| MG-116955 | III | 1991 | C | + | + | E |
| MG-116926 | III | 1991 | C | + | + | E, C |
| MG-117086 | III | 1991 | C | + | + | E |
| MG-117159 | III | 1991 | C | + | + | E |
| MH-08 | III | 1992 | C | + | + | E |
| MAK 757 (El Tor) | Ref | 1937 | E | + | + | E |
| 154 (classical) | Ref | UK | C | + | + | C |
C, classical type; E, El Tor type; Ref, reference strain; UK, unknown; +, positive.
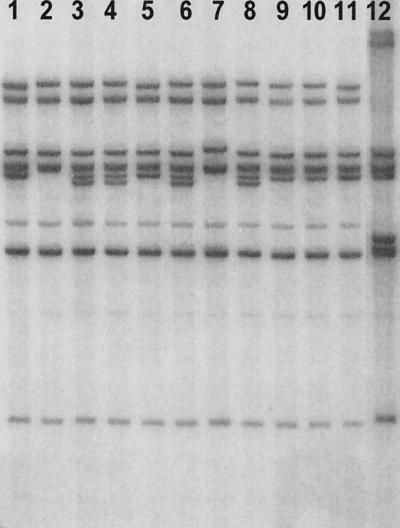
The ribotypes of the V. cholerae strains examined in this study, compared to those of selected reference strains of the El Tor and classical biotypes, are shown in Fig. 1. The ribotypes of different strains representing the three Matlab types of V. cholerae were similar to the ribotypes of El Tor biotype strains and different from that of typical classical biotype strains described previously (11, 12). The ribotypes of two type I strains (lanes 9 and 10) were similar to that of toxigenic El Tor strains 1849 (lane 11), isolated in 2001, and G-3669 (lane 1), isolated in 1969 in Bangladesh. The Matlab type III strains belonged to three different ribotypes (Fig. 1, lanes 2 through 7), and the single type II strain had the same ribotype as a type III strain.
FIG. 1.
BglI restriction patterns of rRNA genes of V. cholerae strains compared to those of selected typical strains of the El Tor and classical biotypes of V. cholerae O1. A Southern blot of BglI-digested genomic DNA was hybridized with the 7.5-kb BamHI fragment of E. coli rRNA clone pKK3535. Lanes (including strain designations and relevant characteristics): 1, toxigenic El Tor strain G-3669 (isolated in 1969 in Bangladesh); 2 through 10, strains MH-08 (Matlab type III), MG-117159 (Matlab type III), MG-117086 (Matlab type III), MG-116926 (Matlab type III), MG-116955 (Matlab type III), MG-116025 (Matlab type III), MG-116226 (Matlab type II), MJ-1485 (Matlab type I), and MJ-1236 (Matlab type I); 11, toxigenic El Tor strain 1849 (isolated in 2001); 12, toxigenic classical biotype strain (isolated in 1963 in Bangladesh).
DISCUSSION
Classical and El Tor strains of V. cholerae are closely related but are not directly derived from each other (16, 17). El Tor vibrios appeared in Bangladesh, causing the first significant outbreak in 1968, and by 1973, they completely replaced the classical vibrios (1). In 1982, the classical biotype reappeared as the predominant epidemic strain in Bangladesh (25). In retrospect, it appears that classical cholera did not completely disappear from Bangladesh during the 1970s or late 1980s, but rather, its frequency varied in different regions of the country (26). The classical and El Tor biotypes have temporally overlapped for over a decade and are likely to have interacted and exchanged genetic material either in the human intestinal milieu or in the aquatic environment. The strains isolated in this study probably represent the amalgam of such an exchange. It is well recognized that genetic exchange between divergent bacterial lineages can contribute importantly to the success of a species in complex and inconstant environments, such as those in which V. cholerae may reside. Several studies have also pointed to such exchanges as an important factor in V. cholerae population genetics and evolution (2, 3, 10).
On the basis of their phenotypic and genotypic traits, Matlab type I strains appeared to be more like the classical biotype while Matlab type II and III strains appeared to be more like the El Tor biotype. Matlab I strains, however, had altered phage receptor sites, since both of the strains were resistant to group IV and V phages. We assessed the similarity of the hybrid strains with classical and El Tor biotype strains on the basis of previously described ribotype patterns of classical and El Tor strains (11, 12). Ribotyping demonstrated that the Matlab I, II, and III strains showed minor differences in fragment patterns shown by the El Tor standard strains, suggesting that the hybrids originated from an El Tor-like clone. Therefore, overall, these strains were of the El Tor biotype displaying traits of the classical biotype. It has been proposed that while El Tor and classical strains are not directly derived from each other but appear to be derived from environmental nontoxigenic strains that are El Tor like (15). Clinical strains might become classical like in some properties simply by loss of function, and this agrees with the finding of the present study. While some genetic exchange has also probably occurred, it appears that the strains have evolved classical biotype properties. With a V. cholerae genomic microarray that displayed more than 93% of the predicted genes of the whole genome sequence of El Tor strain N16961, Dziejman et al. (9) showed that only seven genes were absent solely in classical strains but present in other strains, leading them to speculate that classical biotype strains may be derived from a primordial environmental strain that was more El Tor like than previously thought. Mitra et al. have previously reported the involvement of bacteriophage PS166 in the acquisition of some classical biotype-specific properties by El Tor strains (22, 23). Insertion of lysogenic phage genomes in the bacterial chromosome leading to the activation or inactivation of certain genes or expression of new phage-encoded genes is a natural phenomenon in the origination of genetic diversity. However, in the present study, the acquisition of classical properties such as classical type tcpA and rstR genes by El Tor vibrios by conversion through phage PS166 seems unlikely. It seems more probable that more than one genetic exchanges were involved in the conversion of these strains. Irrespective of the mechanism involved in the generation of the natural hybrid strains, the existence of strains showing a combination of classical and El Tor biotype properties has evolutionary and epidemiological importance.
Interestingly, all of the hybrid strains carried the tcpA gene of the classical type. Recently, the dominance of the classical type tcpA gene among environmental strains of V. cholerae has been reported (6). The primary structure of TcpA is highly conserved among V. cholerae serogroups and biotypes shown to be pathogenic to humans, with amino acid identities of nearly 100% between strains of a given biotype and about 80% between classical and El Tor biotype O1 strains (20). We are not certain if El Tor strains with classical tcpA are more efficient colonizers, but there is enough evidence showing that classical biotype strains elaborate abundant amounts of toxin-coregulated pilin when grown in vitro, in contrast to El Tor strains (20, 29). The strains analyzed in the present study are not only of academic interest but may well represent precursors of other clones that could lead to a pandemic spread since they have all of the genetic features needed to make a V. cholerae strain pandemic. Moreover, these strains were isolated from clinical cases of acute diarrhea. These strains also represent unique natural recombinants that could be judiciously employed in the construction of live-vaccine strains since they have a combination of virulence attributes of both the classical and El Tor biotypes of V. cholerae O1.
The classical biotype of V. cholerae O1 is believed to be extinct and has not been isolated in the past several years, even in southern Bangladesh, the last of the niches where this biotype prevailed (A. K. Siddique, unpublished data). In this study, we show the existence of El Tor strains that have lost some of the El Tor phenotypes and acquired classical biotype characteristics. Therefore, even though strains that represent the classical biotype in entirety have been completely displaced, a reservoir of the virulence genes of the classical biotype still exists in nature. Previous molecular analyses of classical strains isolated between 1961 and 1992 in Bangladesh support the contention that classical vibrios were never completely replaced in Bangladesh (11) Thus, a vaccine developed against cholera must take this into consideration and must be targeted against both of the biotypes, failing which the global use of a vaccine exclusively against the El Tor biotype might select against El Tor strains and favor strains carrying the classical attributes, such as those isolated in this study.
These hybrid strains of V. cholerae may be more common than currently recognized because phenotypic methods are inadequate to precisely distinguish between the two biotypes and are not routinely used in clinical microbiology laboratories. IS1004 fingerprinting has determined that an O37 strain of V. cholerae that was responsible for a large outbreak of cholera in Sudan in 1968 (32) is closely related to classical O1 strains (4). This indicates that horizontal exchange of genes has occurred not only between O1 biotypes but also between classical biotype and non-O1 strains, and the Sudan strain is a typical example of how a novel genotype can cause a large outbreak. Although the strains characterized in this study were isolated a decade ago, the inadequacy of phenotypic tests did not permit us to place these strains accurately. However, molecular techniques have revealed that these strains carry traits of the two biotypes and that such strains exist in nature and are associated with sporadic diarrhea in Bangladesh and, presumably, other areas of the world where cholera is endemic. The recognition of such strains and tracking of their global prevalence and spread are important because each of these types possesses all of the traits necessary to initiate a pandemic spread.
Acknowledgments
This research was funded by a grant from the National Institute of Allergy and Infectious Diseases (grant R01 AI39129); by a grant from the Government of Japan to the Laboratory Science Division of the International Centre for Diarrhoeal Disease Research, Bangladesh; by the Centre for Health and Population Research; and by other core donors of the Centre who share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, the Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States.
We thank Matthew Waldor, New England Medical Center, Boston, Mass., for the rstR gene probes.
REFERENCES
- 1.Bart, K. J., Z. Huq, M. Khan, and W. H. Mosley. 1970. Seroepidemiologic studies during a simultaneous epidemic of infection with El Tor Ogawa and classical Inaba Vibrio cholerae. J. Infect. Dis. 121(Suppl.):S17-S24. [DOI] [PubMed]
- 2.Beltran, P., G. Delgado, A. Davarro, F. Trujillo, R. K. Selander, and A. Cravioto. 1999. Genetic diversity and population structure of Vibrio cholerae. J. Clin. Microbiol. 37:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bik, E. M., A. E Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmid containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, et al. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cholera Working Group. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. [PubMed] [Google Scholar]
- 8.Clemens, J. D., J. R. Harris, D. A. Sack, et al. 1988. Field trial of oral cholera vaccines in Bangladesh: results of one year of follow-up. J. Infect. Dis. 158:60-69. [DOI] [PubMed] [Google Scholar]
- 9.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., M. J. Albert, J. J. Mekalanos. 1998. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., A. R. M. A. Alim, M. M. Rahman, A. K. Siddique, R. B. Sack, and M. J. Albert. 1993. Clonal relationships among classical Vibrio cholerae O1 strains isolated between 1961 and 1992 in Bangladesh. J. Clin. Microbiol. 31:2513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., M. N. Saha, Asadulghani, D. A. Sack, R. B. Sack, Y. Takeda, and G. B. Nair. 2000. The O139 serogroup of Vibrio cholerae comprises diverse clones of epidemic and nonepidemic strains derived from multiple V. cholerae O1 and non-O1 progenitors. J. Infect. Dis. 182:1161-1168. [DOI] [PubMed] [Google Scholar]
- 13.Faruque, S. M., A. K. Siddique, M. N. Saha, et al. 1999. Molecular characterization of a new ribotype of Vibrio cholerae O139 Bengal associated with an outbreak of cholera in Bangladesh. J. Clin. Microbiol. 37:1313-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg, A., and B. Vogelstein. 1984. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 15.Karaolis, D. K., R. Lan, and P. R. Reeves. 1994. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J. Bacteriol. 176:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaolis, D. K. R., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaolis, D. K. R., R. Lan, J. B. Kaper, and P. R. Reeves. 2001. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect. Immun. 69:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keasler, S. P., and R. H. Hall. 1993. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341:1661. [DOI] [PubMed] [Google Scholar]
- 19.Kimsey, H. H., and M. K. Waldor. 1998. CTXφ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirn, T. J., M. J. Lafferty, C. M. P. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 21.Manning, P. A., U. H. Stroeher, and R. Morona. 1994. Molecular basis for O-antigen biosynthesis in Vibrio cholerae: Ogawa-Inaba switching, p. 77-94. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington D.C.
- 22.Mitra, S. N. 1989. Mutation induced by vibriophage Ps166 infection changes biotype and phage type of Vibrio cholerae. J. Med. Microbiol. 30:137-141. [DOI] [PubMed] [Google Scholar]
- 23.Mitra, S. N., R. Mukhopadhay, A. N. Ghosh, and R. K. Ghosh. 2000. Conversion of Vibrio El Tor MAK757 to classical biotype: role of phage PS166. Virology 273:36-43. [DOI] [PubMed] [Google Scholar]
- 24.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, et al. 1994. Spread of Vibrio cholerae O139 Bengal in India. J. Infect. Dis. 169:1029-1034. [DOI] [PubMed] [Google Scholar]
- 25.Samadi, A. R., N. Shahid, A. Eusuf, M. Yunus, M. I. Huq, M. U. Khan, A. S. M. M. Rahman, and A. S. G. Faruque. 1983. Classical Vibrio cholerae biotype displaces El Tor in Bangladesh. Lancet i:805-807. [DOI] [PubMed]
- 26.Siddique, A. K., A. H. Baqui, A. Eusof, K. Haider, M. A. Hossain, I. Bashir, and K. Zaman. 1975. Survival of classic cholera in Bangladesh. Lancet 337:1125-1127. [DOI] [PubMed] [Google Scholar]
- 27.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 28.Stroeher, U. H., K. E. Jedani, B. K. Dredge, et al. 1995. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc. Natl. Acad. Sci. USA 92:10374-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voss, E., and S. R. Attridge. 1993. In vitro production of toxin-coregulated pili by Vibrio cholerae El Tor. Microb. Pathog. 15:255-268. [DOI] [PubMed] [Google Scholar]
- 30.Wachsmuth, K., O. Olsvik, G. M. Evins, and T. Popovic. 1994. Molecular epidemiology of cholera, p. 357-370. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington D.C.
- 31.Yamasaki, S., S. Garg, G. B. Nair, and Y. Takeda. 1999. Distribution of Vibrio cholerae O1 antigen biosynthesis genes among O139 and other non-O1 serogroups of Vibrio cholerae. FEMS Microbiol. Lett. 179:115-121. [DOI] [PubMed] [Google Scholar]
- 32.Zinnaka, Y., and C. C. Carpenter, Jr. 1972. An enterotoxin produced by noncholera vibrios. Johns Hopkins Med. J. 131:403-411. [PubMed] [Google Scholar]