Abstract
We analyzed a collection of 97 well-characterized Burkholderia cepacia genomovar III isolates to evaluate multiple genomic typing systems, including pulsed-field gel electrophoresis (PFGE), BOX-PCR fingerprinting and random amplified polymorphic DNA (RAPD) typing. The typeability, reproducibility, and discriminatory power of these techniques were evaluated, and the results were compared to each other and to data obtained in previous studies by using multilocus restriction typing (MLRT). All methods showed excellent typeability. PFGE with SpeI was more reproducible than RAPD and BOX-PCR fingerprinting. The discriminatory power of the methods was variable, with PFGE and RAPD typing having a higher index of discrimination than BOX-PCR fingerprinting. In general, the results obtained by PFGE, BOX-PCR fingerprinting, and MLRT were in good agreement. Our data indicate that different genomic-based methods can be used to type B. cepacia genomovar III isolates depending on the situation and the epidemiologic question being addressed. PFGE and RAPD fingerprinting are best suited to addressing small-scale studies (i.e., local epidemiology), whereas BOX-PCR fingerprinting is more appropriate for large-scale studies (i.e., global epidemiology). In this regard, BOX-PCR fingerprinting can be considered a rapid and easy alternative to MLRT.
Cystic fibrosis (CF) is the most common hereditary disease in Caucasian populations. Clinical manifestations of CF result from a disturbance in electrolyte transport that primarily affects the respiratory and digestive systems. The CF lung is particularly susceptible to infection with a variety of opportunistic bacteria (9, 14), and exacerbations of chronic infection cause significant morbidity and mortality (36). Among the bacterial species capable of causing infection in CF are those belonging to the Burkholderia cepacia complex, which is currently comprised of nine closely related genomic species or genomovars (11, 21, 45). Recent work has demonstrated that the majority of infected CF patients harbor either B. cepacia genomovar III or Burkholderia multivorans (genomovar II) (1, 25, 38). Furthermore, limited data suggest that B. cepacia genomovar III (or perhaps certain specific strains within genomovar III) may be relatively more virulent than other species in this complex (3, 12, 19).
The broad-spectrum antimicrobial resistance, absence of a vaccine, and virulence of certain strains have made prevention of B. cepacia complex infection an important goal in CF patient care (21, 22). However, much still remains unknown regarding the epidemiology of infection in CF. A number of previous studies have demonstrated transmission of B. cepacia complex strains between persons with CF (for reviews, see references 15, 16, 21, and 22). More recent studies indicate that the natural environment is also a likely reservoir for acquisition of B. cepacia complex strains (4, 24). Better risk assessment of potential sources of infection and the development of optimal infection control policies rely on a more complete understanding of the molecular epidemiology of B. cepacia complex infection in CF.
A number of methods have been used to establish relationships between B. cepacia complex isolates, including phenotypic assays, such as serotyping, antimicrobial susceptibility typing, bacteriocin typing, and biotyping (33, 34). In recent years, phenotypic methods have been largely replaced by genotypic methods, including macrorestriction digestion of chromosomal DNA followed by pulsed-field gel electrophoresis (PFGE) and various PCR-based fingerprinting techniques (2, 31, 43, 44, 48). Among these, PFGE is generally considered the “gold standard” in bacteriological typing (2, 31, 40), and a number of studies have applied PFGE in studies assessing B. cepacia complex epidemiology (1, 7, 10, 37, 46). PCR-based fingerprinting with short random primers (31, 32, 42) or primers directed against repetitive sequences in the bacterial genome (31, 35, 42, 48) are also increasingly being used for typing B. cepacia complex organisms (5, 7, 26, 28-30, 37). Recently, we introduced multilocus restriction typing (MLRT) as yet another method for genotyping B. cepacia complex (10). In MLRT, genomic diversity is indexed through restriction fragment length polymorphism analysis of several housekeeping genes. MLRT is particularly well suited to studies analyzing B. cepacia complex isolates collected in large-scale epidemiologic studies (10).
In the present study, we compared results obtained with PFGE and repetitive sequence PCR by using a BOX A1R primer (BOX-PCR fingerprinting) for a set of 97 epidemiologically well-characterized B. cepacia genomovar III isolates. A subset of these 97 isolates was also analyzed by using random amplified polymorphic DNA (RAPD) typing. Typeability, reproducibility, and discriminatory power of all techniques were compared to each other and to MLRT data obtained in previous studies.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and species identification.
Isolates were obtained from the B. cepacia Research Laboratory and Repository (University of Michigan, Ann Arbor, Mich.) (Table 1) and had been identified as B. cepacia genomovar III by using ribosomal DNA (rDNA)- and recA-based PCR assays, as previously described (23, 27). Approximately two-thirds (n = 66) were recovered from CF sputum culture from persons receiving care in 23 CF treatment centers throughout North America; the rest of the isolates were recovered from soil. All had been typed previously by one or more genotyping methods, including PFGE or MLRT (10). Several clusters of isolates from patients receiving care in the same geographic region were included in this set. Isolates from frozen stocks were grown aerobically on Mueller-Hinton broth (Becton Dickinson) supplemented with 2.2% (wt/vol) agar and incubated overnight at 32°C.
TABLE 1.
List of isolates used and their molecular profiles
| Strain designation | Molecular profile by:a
|
|||
|---|---|---|---|---|
| PFGE (65% similarity) | Box (70% similarity) | MLRTc
|
||
| CC | RT | |||
| AU2718 | P6 | B1 | 6 | 3 |
| AU0199 | P6 | B2 | 6 | 4 |
| AU0604, AU0824 | s | B1 | 6 | 6 |
| AU2105 | s | B1 | 6 | 7 |
| AU2107 | s | B1 | 6 | 8 |
| AU1742 | s | B1 | 6 | 11 |
| HI2812, HI2814, HI2808, HI2824, HI2817, HI2816, HI2815, HI2811, HI2813, HI2825 | P5 | B3 | 6 | 17 |
| AU2567 | P8 | B1 | 6 | 21 |
| AU2638, AU1528, AU1529 | P8 | B1 | 6 | 23 |
| AU2091 | P8 | B1 | 6 | 24 |
| AU1443, AU1472 | P8 | B1 | 6 | 25 |
| AU0067 | P8 | B1 | 6 | 26 |
| AU0916 | s | B1 | 6 | 31 |
| HI2227 | s | B2 | 6 | 146 |
| AU2676, AU0659, AU0660, PC184, AU0915, AU0918, AU2589, AU0089, AU2632, AU0897, AU1526, AU0065, AU0339 | P10 | B8 | 46 | 46 |
| AU2697, AU2689, AU2730, AU0603 | P10 | B8 | 46 | 50 |
| AU0551 | P10 | B8 | 46 | 51 |
| AU2651, AU2725 | P7 | B6 | 71 | 72 |
| AU0787 | s | s | — | 75 |
| HI2766 | s | B4 | — | 76 |
| HI2770 | s | s | 77 | 77 |
| HI2761 | s | B4 | 77 | 78 |
| HI2617 | s | s | — | 79 |
| HI2490 | s | s | — | 80 |
| AU0079 | s | s | 88 | 66 |
| AU2622 | s | s | 88 | 88 |
| AU0137, AU0583, AU0566 | P1 | B5 | 88 | 88 |
| AU0202, PC8, HI2424, | P1 | B5 | 88 | 89 |
| HI2699, HI2566, HI2429 | P2 | B5 | 88 | 89 |
| AU2431, HI2426 | P4 | B5 | 88 | 89 |
| HI2565 | s | B5 | 88 | 89 |
| AU1217 | P1 | s | 88 | 89 |
| AU1107 | P1 | B5 | 88 | 90 |
| AU1482 | P1 | B5 | 88 | 91 |
| HI2571, HI2628, HI2670, HI2677, HI2679 | P2 | B5 | 88 | 99 |
| AU1547 | P3 | B5 | 88 | 109 |
| AU2079 | P3 | B5 | 88 | 112 |
| AU2027 | P3 | B5 | 88 | 114 |
| HI2692 | P4 | B5 | 88 | 118 |
| HI2576, HI2632 | P4 | B5 | 88 | 119 |
| HI2555, HI2558, HI2431, HI2577, HI2689, HI2697, HI2691, HI2683 | P4 | B5 | 88 | 120 |
| HI2485 | s | s | — | 129 |
| AU0208 | s | s | — | 130 |
| AU0670 | P9 | B7 | 132 | 132 |
| AU1644 | P9 | B7 | 132 | 133 |
| ES1405 | s | s | — | 137 |
| ES0263 | s | s | 138 | 138 |
| ES0222 | s | s | 141 | 141 |
| AU0475 | s | s | — | 147 |
Shown are clusters delineated among the profiles with PFGE with a cutoff value of 65% similarity or among the profiles obtained with BOX-PCR with a cutoff value of 70% similarity. MLRT data were taken from previous studies (10; T. Coenye and J. J. LiPuma, submitted for publication). CC, clonal complex; s, separate position in the dendrogram; —, does not belong to a clonal complex (singleton type).
Macrorestriction digest and pulsed field gel electrophoresis.
Single bacterial colonies were removed from an agar plate, suspended in 1 ml of SE buffer (75 mM NaCl, 25 mM EDTA [pH 7.4]), pelleted by centrifugation at 4,300 × g for 3 min, washed 3 times in 1 ml of SE buffer, and resuspended in 0.5 ml of SE buffer. The optical density at 620 nm was adjusted to approximately 1.0. Two hundred microliters of the cell suspension was homogenized with 200 μl of 2% low-melting-temperature agarose (Sigma, St Louis, Mo.) in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA) and poured into a plug mold. After 15 min at 4°C, agarose plugs were placed in 10 ml of PEN buffer (1.0% N-lauryl sarcosine, 500 mM EDTA [pH 9.6]) containing 1 mg of protease ml−1. After incubation at 37°C in a rocking incubator for 12 to 18 h, plugs were washed four times for 1 h per wash with 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). Two-millimeter-wide plug sections were cut and incubated for 4 h with 5 U of SpeI (Promega, Madison, Wis.) in 115 μl of digestion buffer. DNA fragments were separated in 1% PFGE-certified agarose (Bio-Rad, Hercules, Calif.) by using a CHEF DRIII system (Bio-Rad). Plugs containing digested DNA of B. cepacia strain AU2725 were included on each gel to allow intra- and intergel normalization. A current of 5.0 V/cm was applied for 25 h, with pulse times of 30 to 70 s (linear ramping). Gels were stained with ethidium bromide and visualized with UV illumination. Gel images were digitized using a GelDoc2000 gel analyzer (Bio-Rad) and stored as TIF files. Digitized images were converted, normalized with the reference lanes (containing DNA from strain AU2725), and analyzed by using Molecular Analyst Fingerprinting Plus software (Bio-Rad). The rolling disk background subtraction method was applied, and similarity matrices of densitometric curves of the gel tracks (the first 16% and last 7.5% of data points were excluded from the analysis) were calculated by using Pearson's product-moment correlation coefficient. Cluster analyses of similarity matrices were performed by the unweighted pair group method with arithmetic averages (UPGMA).
BOX-PCR fingerprinting.
DNA from each isolate was prepared by heating one colony at 95°C for 15 min in 20 μl of lysis buffer containing 0.25% (wt/vol) sodium dodecyl sulfate (SDS) and 0.05 M NaOH. Following lysis, 180 μl of distilled water was added, and the DNA solutions were stored at 4°C. Rep-PCR typing with a BOX-A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) (BOX-PCR fingerprinting) was carried out as described previously (35). Briefly, 2 μl of DNA solution was mixed with 2 U of Taq polymerase (Gibco BRL, Gaithersburg, Md.), 1.25 μl of 25 mM (each) of deoxynucleotide triphosphate (Gibco BRL), 2.5 μl of dimethyl sulfoxide (DMSO), 0.4 μl of bovine serum albumin (20 mg ml−1) (Promega), 5 μl of 5× Gitschier buffer and 1 μl of primer (0.3 μg μl−1) in a final volume of 25 μl. Amplification was carried out with a PTC-100 programmable thermal cycler (MJ Research, Incline Village, Nev.). After initial denaturation for 2 min at 95°C, 35 amplification cycles were completed, each consisting of 3 s at 94°C, 30 s at 92°C, 1 min at 50°C, and 8 min at 65°C. A final extension of 8 min at 65°C was applied. PCR products were separated on 25-cm-long 1.5% agarose gels in 0.5× TBE buffer (60 mA for 4 h at room temperature). A 1-kb molecular weight marker (Gibco) was used multiple times on each gel to allow normalization. Following staining with ethidium bromide and visualization by UV illumination, gels were analyzed as described above for PFGE (the first 12.5% and last 8.5% of data points were excluded from the analysis).
RAPD typing.
DNA was prepared as described above for BOX-PCR. RAPD fingerprinting was performed with primer RAPD-270 (5′-TGCGCGCGGG-3′) as described previously (7, 28). Briefly, 2 μl of DNA solution was mixed with 1 U of Taq polymerase (Gibco BRL, Gaithersburg, Md.), 2.5 μl of 2.5 mM (each) deoxynucleotide triphosphates (Gibco BRL), 0.7 μl of bovine serum albumin (20 mg ml−1) (Promega), 2.5 μl of 10× buffer (100 mM Tris-HCl [pH 8.0], 0.5 M KCl, 30 mM MgCl2, 1.0% gelatin), and 0.4 μl of primer (100 pmol μl−1) in a final volume of 25 μl. Amplification was carried out with a RapidCycler programmable thermal cycler (Idaho Tech, Idaho Falls, Idaho). The first four amplification cycles were each for 1 min at 94°C, 1 min at 36°C, and 2.5 min at 72°C. The following 29 cycles were 30 s at 94°C, 30 s at 36°C, and 75 s at 72°C. A final extension of 2 min at 72°C was applied. Numerical analysis was performed as described above for PFGE (the first 15.75% of data points were excluded from the analysis).
Statistical analyses.
Two-tailed unpaired t tests were used to compare reproducibility among replicate assays within a given typing method and were calculated by using GraphPad Prism 3.00 (GraphPad Software, San Diego, Calif.). Discriminatory index (DI) is the probability that two isolates randomly chosen from a population of unrelated isolates will be distinguished by a given typing method within the confines of a specific cutoff value. DI is determined by the number and relative frequencies of the different types defined by a given method and is calculated by using Simpson's index of diversity (17) as follows:
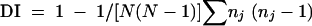 |
where N is the total number of isolates and nj is the number of isolates belonging to the jth type. The cophenetic correlation coefficient is the product-moment correlation between all original matrix similarities and all corresponding similarity values derived from the resulting dendrogram. As such, it provides a measure of whether or not a matrix can be faithfully represented as a bifurcating tree. Cophenetic correlation coefficients of the UPGMA dendrograms produced by using each typing method were calculated by using Molecular Analyst software.
RESULTS
PFGE typing.
An illustration of PFGE patterns is shown in Fig 1. Reproducibility was checked by preparing agarose-embedded DNA from 15 isolates two or more times. Similarity coefficients between these replicates ranged between 90.7 and 97.6% (mean ± standard deviation, 93.84% ± 2.34%). Among the 97 isolates studied, 10 clusters could be delineated (designated P1 to P10) by using a cutoff value of 65% similarity, while 22 isolates occupied separate positions in the dendrogram (Fig. 2 and Table 1). Isolates belonging to the same cluster did not differ by more than six bands, although in general, isolates belonging to different clusters differed by more than six bands (data not shown). The DI for PFGE by using a 65% similarity cutoff value was 0.920. The cophenetic correlation coefficient of the UPGMA dendrogram was 79.7%.
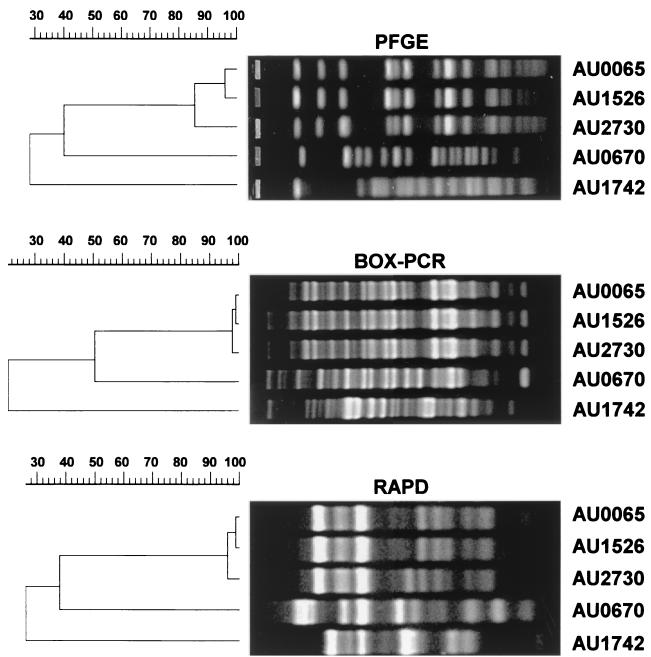
FIG. 1.
Illustration of genotyping methods with representative B. cepacia complex genomovar III isolates. PFGE, BOX-PCR, RAPD gels, and UPGMA dendrograms were produced as described in the text. Scale bars indicate percent similarity.
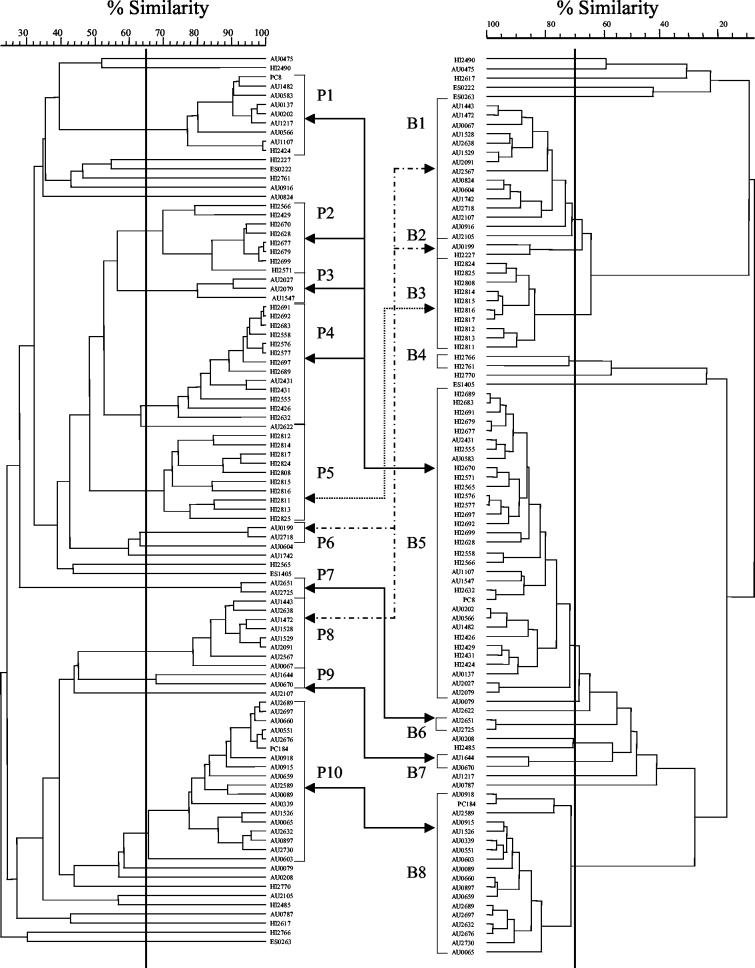
FIG. 2.
Dendrograms derived from the UPGMA linkage of correlation coefficients between the PFGE patterns (left) and BOX-PCR patterns (right). Clusters were delineated with a 65% similarity cutoff for PFGE and a 70% similarity cutoff level for BOX-PCR, as indicated by the heavy vertical lines. Corresponding clusters are indicated by connecting arrows. (Solid, dashed, or dotted lines are included for ease of interpretation only.)
BOX-PCR fingerprinting.
An illustration of BOX-PCR patterns is shown in Fig. 1. Reproducibility was assessed by obtaining a pattern from 15 isolates two or more times. Similarity indices between these replicates ranged between 83.6 and 97.2% (mean ± standard deviation, 90.27% ± 3.88%). By using a cutoff value of 70% similarity, eight clusters could be delineated (designated B1 to B8), while 13 isolates occupied separate positions in the dendrogram (Fig. 2 and Table 1). The DI of BOX-PCR fingerprinting with a 70% similarity cutoff value was 0.821. The cophenetic correlation coefficient of the dendrogram was 96.3%.
RAPD typing.
From the same set of 97 isolates analyzed by PFGE and BOX-PCR, a subset of 31 isolates (recovered from CF patients receiving care in four different treatment centers in the same U.S. state) was further investigated by using RAPD typing. An illustration of RAPD typing is shown in Fig. 1. Reproducibility was assessed by obtaining a pattern from six isolates two times. Similarity indices between these replicates ranged from 71.0 to 97.4% (mean ± standard deviation, 83.88% ± 10.39%). With a cutoff value of 80% similarity, three clusters could be delineated (designated R1 to R3), while six isolates occupied separate positions in the dendrogram (Fig. 3 and Table 2). The DI for RAPD typing with an 80% similarity cutoff value was 0.738.
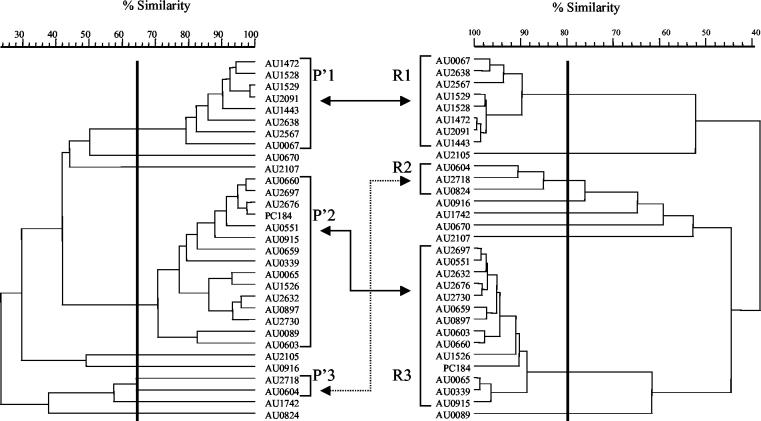
FIG. 3.
Dendrograms derived from the UPGMA linkage of correlation coefficients between the PFGE patterns (left) and RAPD patterns (right). Clusters were delineated with 65 and 80% similarity cutoff values for PFGE and RAPD, respectively, as indicated by the heavy vertical lines.
TABLE 2.
Subset of isolates investigated by RAPD typing
| Strain designation | Molecular profile bya:
|
|
|---|---|---|
| PFGE (65% similarity) | RAPD (80% similarity) | |
| AU2567, AU2638, AU1528, AU1529, AU2091, AU1443, AU1472, AU0067 | P′1 | R1 |
| AU2676, AU0659, AU0660, AU2697, AU0551, PC184, AU0915, AU0089, AU0603, AU2632, AU0897, AU1526, AU2730, AU0065, AU0339 | P′2 | R3 |
| AU2718, AU0604 | P′3 | R2 |
| AU0824 | s | R2 |
| AU2105, AU2107, AU1742, AU0670, AU0916 | s | s |
Shown are clusters delineated among the profiles obtained with PFGE with a cutoff value of 65% similarity or among the profiles obtained with RAPD with a cutoff value of 80% similarity. s, separate position in the dendrogram.
Comparison of methods.
We found a good correlation between PFGE and BOX-PCR fingerprinting. Most clusters delineated in the dendrogram based on BOX-PCR fingerprinting with a 70% coefficient cutoff have corresponding clusters in the PFGE dendrogram with a 65% cutoff (Fig. 2 and Table 1). Specifically, isolates grouping in clusters P5, P7, P9, and P10 in the PFGE dendrogram group in clusters B3, B6, B7, and B8, respectively, in the BOX-PCR dendrogram. The 10 isolates in cluster P5/B3 were recovered from patients receiving care in nine Canadian cities; these had previously been identified by PFGE analysis as ET12, the genomovar III strain that predominates among infected CF patients in Canada and the United Kingdom (18). The 18 isolates in cluster P10/B8 were recovered from CF patients receiving care in the same U.S. state.
Isolates in PFGE clusters P1, P2, P3, and P4 group together in the BOX-PCR dendrogram in cluster B5. The isolates in cluster P1 were recovered from patients in two large CF treatment centers in the same region of the U.S. (7), while those in P2 and P4 were recovered from soil in the same geographic region (24).
Isolates belonging to BOX-PCR cluster B1 either group in cluster P6 or P8 or occupy separate positions in the dendrogram based on PFGE. The 10 isolates in clusters P6 and P8 were recovered from patients in the same U.S. state. Cluster B2 is composed of one isolate belonging to cluster P6 and one isolate that occupies a separate position in the PFGE dendrogram. BOX-PCR cluster B4 is composed of two isolates that occupy separate positions in the PFGE dendrogram.
In general, there was also good agreement between PFGE and RAPD typing for the subset of 31 isolates (Fig. 3 and Table 2). Isolates grouping in clusters P′1 and P′2 in the PFGE dendrogram group in clusters R1 and R3, respectively, in the RAPD dendrogram with a cutoff of 80%. Isolates AU2718 and AU0604 group together in PFGE cluster P′3; these isolates group together with isolate AU0824 in RAPD cluster R3. The discriminatory powers of PFGE (DI = 0.712) and RAPD (DI = 0.738) were similar for this data set.
We also compared the results obtained with BOX-PCR and PFGE with those previously obtained with MLRT analysis of the same set of 97 isolates (10; and T. Coenye and J. J. LiPuma, submitted for publication). MLRT allows the grouping of isolates in a hierarchical fashion such that isolates are first grouped by restriction type (RT), consisting of a unique combination of RFLP profiles for five genetic loci. RTs are then grouped into clonal complexes, defined as groups in which each RT is identical to at least one other RT at three or more of the five loci (T. Coenye and J. J. LiPuma, submitted for publication). For the data set used in this study, the DI of MLRT is 0.952 when using the RT as a cutoff; the DI is considerably lower (0.756) when the cutoff is placed at the clonal complex level. As reported previously (10), there was a good correlation between data obtained with MLRT and data obtained with PFGE (Table 1). In the present analysis, we also found an excellent correlation between data obtained with MLRT and that obtained with BOX-PCR fingerprinting (Table 1); overall, 95.9% of isolates were grouped the same by both methods.
DISCUSSION
Previous studies regarding the molecular epidemiology of infection due to B. cepacia complex species have employed a variety of genotyping methods, including ribotyping, PFGE, RAPD, ERIC-PCR, and BOX-PCR fingerprinting (5, 7, 10, 24, 26, 28, 29, 30, 37, 46). Among these methods, PFGE and RAPD have emerged as the most widely used in recent studies; however, few studies have compared different genotyping methods in a systematic way. Bingen et al. (5) concluded that RAPD was less discriminative than PFGE for the study of bacteria in the B. cepacia complex. In contrast, Liu et al. (26) showed that these methods had comparable discriminatory power, but noted that PFGE was considerably more reproducible. These investigators also concluded that ERIC-PCR should be considered a valid and reproducible alternative to PFGE. However, in both studies, relatively small sets of B. cepacia complex isolates were evaluated in the comparison of these methods. In a more recent study, we explored the use of MLRT as an alternative genotyping method and showed a strong correlation between PFGE and MLRT in an analysis of a larger set of B. cepacia genomovar III isolates (10). In the present study, we sought to expand these findings to include other commonly used PCR-based methods, including BOX-PCR and RAPD typing. We also wished to more specifically identify the strengths and weaknesses of these methods for typing B. cepacia genomovar III, to compare these results with MLRT, and to determine which methods are best suited for addressing questions in the settings of local and global epidemiology.
Several criteria have been used in evaluating bacterial genotyping methods, including typeability, reproducibility, discriminatory power, and ease of interpretation (2, 20, 31). Typeability describes the ability of a given method to provide a readable result for each isolate analyzed (2, 6). All isolates included in this study were typeable by each method used. In previous work, we have noted that a small fraction of B. cepacia complex isolates are refractory to PFGE typing, presumably due to the presence of high DNase activity (data not shown).
Reproducibility measures the ability of a technique to yield the same result when replicate assays are performed on the same isolate (2). In this study, we found that PFGE was significantly more reproducible than BOX-PCR and RAPD typing (P < 0.05). The reproducibility values of PFGE were also in a significantly narrower range than those of BOX-PCR (P < 0.05) and RAPD (P < 0.0001). The high reproducibility of PFGE is in agreement with findings of most previous studies (2, 5, 31, 37, 40). In contrast, previous studies have provided conflicting data regarding the reproducibility of RAPD typing (2, 5, 26, 28, 31, 32, 37, 41, 42). Excellent reproducibility was reported by Mahenthiralingam et al. (28) and Segonds et al. (37), while significant day-to-day variation in RAPD patterns obtained from the same B. cepacia complex isolate was noted by Bingen et al. (5) and Liu et al. (26). In our study, we similarly found the reproducibility of RAPD profiles to be rather low. Reproducibility of BOX-PCR fingerprinting was intermediate between that of PFGE and RAPD; moreover, the values we obtained were generally in agreement with those reported by others (8, 35).
Discriminatory power defines the ability of a typing method to distinguish different strains. This may be expressed as an index that measures the probability that two unrelated strains will be placed into different groups. Discriminatory power is most conveniently calculated by using Simpson's index of diversity (DI), which takes into account the number of types defined by the method and the relative frequencies of these types (2, 17). A DI value of >0.90 has been considered adequate in previous assessments of genotyping methods (13, 17, 39, 47). In our study, only PFGE (with DI = 0.920, with this data set and a 65% similarity coefficient cutoff) would meet this stringent criterion. However, higher discriminatory power does not necessarily always result in a more accurate representation of epidemiologic relatedness. While this may be true in the setting of outbreak epidemiology, in which very high discriminatory power is needed to trace patient-to-patient transmission or nosocomial outbreaks, methods based on such a high DI may very well be less suitable for the analysis of large populations of organisms collected over extended periods of time. This is illustrated by the results previously obtained by using MLRT analysis of this same set of B. cepacia genomovar III isolates. Important relationships among isolates grouped at the clonal complex level (i.e., employing a significantly lower DI) are not apparent based on PFGE analysis. In this regard, MLRT provides greater utility in allowing meaningful analysis of relationships among isolates collected both in larger and smaller scale (both temporal and spatial) studies (10). From this perspective, it is important to note that, in this study, the DI of BOX-PCR fingerprinting with a 70% similarity cutoff (DI = 0.821) is intermediate between the DIs of MLRT analysis according to either RT (DI = 0.952) or clonal complex (DI = 0.756)-level cutoffs. Of course, the discriminatory power of any given typing method is determined by the data set and by the cutoff values used. The cutoff values used in this study were based on (i) the available epidemiologic data (i.e., geographic location and previously determined epidemiological relationships), (ii) the reproducibility of the methods, and (iii) the cophenetic correlation coefficient. It is possible that with other data sets, these similarity coefficient cutoff values may need to be adjusted to better accommodate these variables.
A final criterion to consider in assessment of typing methods is ease of performance. This includes not only performance of the assay, but also interpretation of the resulting data. PFGE is without doubt the more laborious and time-consuming technique among those examined. In contrast to PCR-based methods, PFGE requires a much longer time to perform (4 days with the protocol described above) and more specialized equipment (2, 31). BOX-PCR and RAPD fingerprinting are significantly less cumbersome; results can be obtained within 1 working day. MLRT occupies an intermediate position in this regard.
Whereas consensus guidelines for interpreting DNA restriction patterns generated by PFGE have been published (40), comparable criteria for interpretation of RAPD and BOX-PCR patterns are not available. However, the PFGE interpretative criteria were intended to be used only as an aid in the visual examination of small sets of isolates related to putative outbreaks of disease. These criteria are impractical and quite limited for analyzing larger sets of isolates where multiple pairwise comparisons are required. Indeed, for all the genotyping methods under consideration, visual comparison of large number of complex fingerprint patterns is not only time-consuming but also highly subjective. The use of equipment to digitize patterns and software to perform numerical analysis of these patterns are necessary for studies involving typing of a significant number of isolates.
The data presented in this study indicate that there are strengths and weaknesses among the various genotyping methods that have been used to investigate the epidemiology of B. cepacia genomovar III. Which technique is chosen for a given study depends not only on the preferences of the investigators and the resources available, but most importantly on the specific epidemiologic question being addressed. Our data indicate that the reproducibility of RAPD typing is not sufficient to allow reliable comparisons across large numbers of assays in large-scale studies. This relatively poor reproducibility also limits the portability of results between laboratories. On the other hand, RAPD analysis is well suited to smaller-scale studies, such as investigation of a hospital outbreak, in which a limited number of samples is collected within a narrow time frame. In this setting, PFGE would be a more reproducible and portable, but also more time-consuming and expensive alternative. For more global epidemiological questions, involving larger number of isolates collected over a longer time frame, both MLRT and BOX-PCR fingerprinting might be considered the methods of choice. While data derived from MLRT can be used for population structure analysis (10), BOX-PCR fingerprinting has the advantage of being a more rapid and less expensive method.
Acknowledgments
This work was supported by a grant from the Cystic Fibrosis Foundation (United States) (to J.J.L.). T.C. is supported by the Caroll Haas Research Fund in Cystic Fibrosis.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Giannino, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit, R. D. 1995. Laboratory procedures for the epidemiologic analysis of microorganisms, p 190-208. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 3.Aris, R. M., J. C. Routh, J. J. LiPuma, D. G. Heath, and P. H. Gilligan. 2001. Lung transplantation for cystic fibrosis patients with Burkholderia cepacia complex: survival linked to genomovar type. Am. J. Respir. Crit. Care Med. 164:2102-2106. [DOI] [PubMed] [Google Scholar]
- 4.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Env. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen, E. H., M. Weber, J. Derelle, N. Brahimi, N. Y. Lambert-Zechovsky, M. Vidailhet, J. Navarro, and J. Elion. 1993. Arbitrarily primed polymerase chain reaction as a rapid method to differentiate crossed from independent Pseudomonas cepacia infections in cystic fibrosis patients. J. Clin. Microbiol. 31:2589-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burucoa, C., V. Lhomme, and J. L. Fauchere. 1999. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis. J. Clin. Microbiol. 37:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. S., K. A. Witzmann, T. Spilker, R. J. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 8.Cho, J.-C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing, a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 11.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Soyza, A., A. McDowell, L. Archer, J. H. Dark, S. J. Elborn, E. Mahenthiralingam, K. Gould, and P. A. Corris. 2001. Burkholderia cepacia complex genomovars and pulmonary transplantation outcomes in patients with cystic fibrosis. Lancet 358:1780-1781. [DOI] [PubMed] [Google Scholar]
- 13.Dillon, J.-A., M. Rahman, and K.-H. Yeung. 1993. Discriminatory power of typing schemes based on Simpson's index of diversity for Neisseria gonorrhoeae. J. Clin. Microbiol. 31:2831-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilligan, P. H. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 4:35-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govan, J. R. W., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographical and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledson, M. J., M. J. Gallagher, M. Jackson, C. A. Hart, and M. J. Walshaw. 2002. Outcome of Burkholderia cepacia colonisation in an adult cystic fibrosis centre. Thorax 57:142-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LiPuma, J. J. 1998. Molecular tools for epidemiologic study of infectious diseases. Pediatr Infect. Dis J 17:667-675. [DOI] [PubMed] [Google Scholar]
- 21.LiPuma, J. J. 1998. Burkholderia cepacia: management issues and new insights. Clin. Chest. Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 22.LiPuma, J. J. 1998. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Cur. Opin. Pulm. Med. 4:337-441. [DOI] [PubMed] [Google Scholar]
- 23.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 25.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 26.Liu, P. Y.-F., Z.-Y. Dhi, Y.-J. Lau, B.-S. Hu, J.-M. Shyr, W.-S. Tsai, Y.-H. Lin, and C.-Y. Tseng. 1995. Comparison of different PCR approaches for characterization of Burkholderia (Pseudomonas) cepacia isolates. J. Clin. Microbiol. 33:3304-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for the identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahenthiralingham, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyawaki, H., J. Fujita, K. Takigawa, K. Negayama, Y. Yamagishi, Y. Yamaji, K. Ouchi, T. Nakazawa, K. Kawanishi, and J. Takahara. 1995. Investigation of nosocomial respiratory infection due to Pseudomonas cepacia by arbitrarily primed polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 23:77-83. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki, M., T. Watanabe, K. Morita, Y. Higurashi, K. Araki, N. Shukuya, S. Baba, N. Watanabe, T. Egami, N. Furuya, M. Kanamori, S. Shimazaki, and H. Uchimura. 1999. Molecular epidemiological investigation using a randomly amplified polymorphic DNA assay of Burkholderia cepacia isolates from nosocomial outbreaks. J. Clin. Microbiol. 37:3809-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Power, E. G. 1996. RAPD typing in microbiology—a technical review. J. Hosp. Infect. 34:247-265. [DOI] [PubMed] [Google Scholar]
- 33.Rabkin, C. S., W. R. Jarvis, R. L. Anderson, J. Govan, J. Klinger, J. LiPuma, W. J. Martone, H. Monteil, C. Richard, S. Shigeta, A. Sosa, T. Stull, J. Swenson, and D. Woods. 1989. Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev. Infect. Dis. 11:600-607. [DOI] [PubMed] [Google Scholar]
- 34.Rabkin, C. S., W. R. Jarvis, and W. J. Martone. 1987. Current status of Pseudomonas cepacia typing systems. Eur. J. Epidemiol. 3:343-346. [DOI] [PubMed] [Google Scholar]
- 35.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterisation of the diversity of ecologically important microbes by rep-PCR fingerprinting, p 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Rosenstein, B. J., and P. L. Zeitlin. 1998. Cystic fibrosis. Lancet 351:277-282. [DOI] [PubMed] [Google Scholar]
- 37.Segonds, C., E. Bingen, G. Couetdic, S. Mathy, N. Brahimy, N. Marty, P. Plesiat, Y. Michel-Briand, and G. Chabanon. 1997. Genotypic analysis of Burkholderia cepacia isolates from 13 French cystic fibrosis centers. J. Clin. Microbiol. 35:2055-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struelens, M. J., and European Study Group on Epidemiological Markers, European Society for Clinical Microbiology and Infectious Diseases. 1996. Consensus guidelines for appropriate use and evaluation of epidemiological typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyler, K. D., G. Wang, S. D. Tyler, and W. M. Johnson. 1997. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 35:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Belkum, A. 1994. DNA fingerprinting of medically important microorganisms by use of PCR. Clin. Microbiol. Rev. 7:174-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Belkum, A., M. Sluijter, R. de Groot, H. Verbrugh, and P. W. Hermans. 1996. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 34:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound test results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Looveren, M., C. A. Ison, M. Ieven, P. Vandamme, I. M. Martin, K. Vermeulen, A. Renton, and H. Goossens. 1999. Evaluation of discriminatory power of typing methods for Neisseria gonorrhoeae. J. Clin. Microbiol. 37:2183-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 25:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]