Abstract
Recently, a novel DNA virus designated SEN virus (SEN-V), which is thought to be related to posttransfusion hepatitis, was discovered. The aim of the present study was to clarify the relationship between SEN-V infection and the development of liver disease. We examined SEN-V from the sera of 21 patients with non-B, non-C hepatocellular carcinoma (HCC) and 13 patients with non-B, non-C chronic liver disease (CLD) without HCC who were admitted to our hospital between 1995 and 1997. Thirty-two patients without liver disease served as controls and were also examined for SEN-V. SEN-V DNA was detected by the nested PCR method after extraction of DNA from serum. SEN-V DNA was detected in 74% (25 of 34) of patients with CLD with or without HCC who were negative for both hepatitis B virus surface antigen and anti-hepatitis C virus antibody. SEN-V DNA was detected in 69% (9 of 13) of CLD patients without HCC and in 76% (16 of 21) of HCC patients. The prevalence of SEN-V was no higher in patients with liver disease than in patients without liver disease (24 of 32; 75%). There were no significant differences in age, sex, liver function, history of blood transfusion, or amount of alcohol intake between SEN-V-positive and SEN-V-negative CLD and HCC patients. Genetic analysis suggested that SEN-V is closely related to the TT virus family. SEN-V was detected at almost the same frequency in patients with and without liver disease. SEN-V does not seem to contribute either to the pathogenesis of liver disease or to the development of HCC from chronic liver disease.
The number of patients with hepatocellular carcinoma (HCC) has been increasing over the last 30 years in Japan, and approximately 30,000 patients die of HCC every year (13, 14). The majority of these patients are positive for either hepatitis B virus surface antigen (HBsAg) or anti-hepatitis C virus (anti-HCV) antibody. In a recent study, 11% of HCC patients were positive for HBsAg and 84% were positive for anti-HCV, but the remaining 4% were negative for both anti-HCV and HBsAg (16). The pathogenic agent in these 4% of patients remains unknown.
An RNA virus associated with non-A to E hepatitis named GB virus C (GBV-C)/hepatitis G virus (HGV) and a DNA virus named TT virus (TTV) were recently cloned (6, 8, 11, 12, 17). However, previous studies showed that GBV-C/HGV and TTV are unlikely to be major etiologic agents of non-B, non-C HCC, and they have not yet been confirmed to be hepatotropic (1-5, 7, 9, 10, 24).
A novel DNA virus named SEN virus (SEN-V) was also recently cloned from the plasma of a patient with posttransfusion hepatitis. This patient had elevated transaminase levels but did not have viral markers for hepatitis type A to E viruses or hepatitis G virus (19). SEN-V is a circular 3,900-nucleotide DNA virus. There are eight SEN-V genotypes, called A to H. SEN-V was classified into the same group as TTV variants (including SANBAN and TUS01). Prototype TTV, TTV YONBAN, and TTV PMV formed an outer group of SEN-V (19). Of the eight genotypes, genotypes D and H (or genotype C) have been reported to be related to liver dysfunction (19). Although SEN-V has been observed in patients with acute and chronic liver disease (CLD) of unknown etiology, the role of SEN-V in the pathogenesis of liver disease is not yet known (15, 21, 22). We screened patients with non-B, non-C CLD and patients without liver disease for SEN-V DNA to determine the possible role of SEN-V in the pathogenesis of liver disease and in the development of HCC, especially in patients with non-B, non-C chronic hepatitis and HCC.
MATERIALS AND METHODS
Patients.
We examined the prevalence of SEN-V DNA in 21 patients admitted to the University of Tokyo Hospital between 1995 and 1997 who were diagnosed with non-B, non-C HCC (the HCC patient group) and in 13 patients with non-B, non-C CLD without HCC (the CLD patient group). Thirty-two patients without liver disease were also examined and served as a control group. These patients presented to our hospital with abdominal complaints but had normal transaminase levels. The diagnosis of HCC was made by ultrasonography, computed tomography, magnetic resonance imaging, angiography, and liver biopsy. The clinical features examined included age, sex, history of blood transfusion, history of alcohol intake, serum transaminase levels, platelet count, and indocyanine green (ICG) retention rate (percent) at 15 min (the ICG R15 test). Anti-HCV was identified by a second-generation enzyme immunoassay (Ortho Diagnostics, Tokyo, Japan), and HBsAg was identified by radioimmunoassay (Abbott Laboratories, North Chicago, Ill.). Patients with an ethanol intake exceeding 80 g/day for more than 10 years were considered to have a positive history for alcohol abuse. Patients who were positive for antinuclear antibody or anti-smooth muscle antibody or who had a positive lupus erythematosus test result were excluded. Patients who had previously been treated with antiviral drugs, such as interferon and lamivudine, were also excluded. Serum samples from all patients with HCC were obtained at the first admission after the diagnosis of HCC. The serum samples were stored at −30°C.
Detection of SEN-V DNA.
DNA was extracted from 50 μl of serum with the SepaGene kit (Sanko Junyaku, Tokyo, Japan) according to the manufacturer's instructions. The extracted DNA was dissolved in 20 μl of Tris-HCl buffer (10 mM; pH 8.0) containing 1 mM EDTA, heated to 95°C for 15 min, and quickly chilled on ice. The total amount of extracted DNA was subjected to nested PCR with Ready-To-Go PCR beads (Pharmacia Biotech, Uppsala, Sweden). The first round of PCR was performed with primer sets specific for each genotype. The sense primers specific for genotypes A to D and H were as follows: SEA-S, SEB-S, SEC-S, SED-S, and SEH-S, respectively (Table 1). The antisense primers specific for genotypes A to D and H were as follows: SEA-AS, SEB-AS, SEC-AS, SED-AS, and SEH-AS, respectively (Table 1). These primers were used to amplify 336-, 444-, 619-, 510-, and 830-bp fragments from genotypes A to D and H, respectively. The reaction was run in microtubes with the GeneAmp PCR system 9600 (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Amplification was performed for 35 cycles, with each cycle consisting of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and elongation at 72°C for 1 min. A 10-min final hold at 72°C was used to complete strand synthesis. The second round of PCR was carried out with sense and antisense primers specific for genotypes A to D and H. The sense primers were as follows: SEA-S2, SEB-S2, SEC-S2, SED-S2, and SEH-S2, respectively (Table 1). The antisense primers specific for genotypes A to D and H were as follows: SEA-AS2, SEB-AS2, SEC-AS2, SED-AS2, and SEH-AS2, respectively (Table 1). These primers were used to amplify 288-, 396-, 577-, 459-, and 783-bp fragments from genotypes A to D and H, respectively. The amplification was performed for 30 cycles, with each cycle consisting of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and elongation at 72°C for 1 min, followed by a final hold at 72°C for 7 min. The amplified products were electrophoresed on 1.5% agarose gels, stained with ethidium bromide, and observed under UV light.
TABLE 1.
Primers used to detect or sequence SEN-V
| Sense primer (sequence) | Antisense primer (sequence) | Region amplified (nucleotides) |
|---|---|---|
| SEA-S (5′-TATAACTAACCGCACTTCC-3′) | SEA-AS (5′-CTAAAGCAGCATTCCCAGT-3′) | 78-413 |
| SEB-S (5′-TATCTGCCTGTTACCTCCT-3′) | SEB-AS (5′-TAAATGCTGCTCTGTTCTCT-3′) | 473-916 |
| SEC-S (5′-TCTACGAGTGCATGTTCCG-3′) | SEC-AS (5′-TCTCTGGTACTGGTCATAG-3′) | 389-1007 |
| SED-S (5′-TATCTGCCTACTGCCAATC-3′) | SED-AS (5′-GGCTGAATGTGACTGTGCT-3′) | 471-980 |
| SEH-S (5′-TGCTATGAGTTGGAGACCC-3′) | SEH-AS (5′-TGAGTGTTAGGGGCTGTGT-3′) | 357-1186 |
| SEA-S2 (5′-GCTGAGTTTTCCACGCC-3′) | SEA-AS2 (5′-CTTTCTCTGCCGGCGG-3′) | 102-389 |
| SEB-S2 (5′-CTGGACCCTTCCTCGG-3′) | SEB-AS2 (5′-ACAGTAGTAGAGGCAGC-3′) | 497-892 |
| SEC-S2 (5′-CACATGCTGCTAGCTGT-3′) | SEC-AS2 (5′-CACTTTGAGAGAGAAAGTC-3′) | 410-986 |
| SED-S2 (5′-GTGGGACCCTCCCAAC-3′) | SED-AS2 (5′-CTCCAAACTTTTCTGGCTG-3′) | 495-953 |
| SEH-S2 (5′-GCACAACCCCAATGGGA-3′) | SEH-AS2 (5′-GTCCATTTTCATTGGGGGA-3′) | 381-1163 |
| SED-S3 (5′-TCCCGCTTGAGACCCCT-3′) | 528 | |
| SEH-S3 (5′-GAGAAACCTGTGGGAGG-3′) | 402 | |
| SED Full F (5′-ACGTCACTAACCACGTGAC-3′) | SED(1) R (5′-AGAGCGGRAAGGGGTCTCAAG-3′) | 1-553 |
| SED(3) F (5′-CCTTATCATGTGCGGTGC-3′) | SED(3) R (5′-GGTTTTGACTTCCATCTGC-3′) | 868-1589 |
| SED(4) F (5′-AACGACATGATAGGCTTCC-3′) | SED(4) R (5′-CCTACTGTCTCGTGGTCTTGAA-3′) | 1439-2082 |
| SED(5) F (5′-TCTACAAAACCTACCTCTGTGG-3′) | SED(5) R (5′-GCTTCTTTTTCTTGGGAGCTG-3′) | 1990-2688 |
| SED(6) F (5′-AGAAAAAGACTCAGGTTCGGA-3′) | SED Full R (5′-CAATGGCAACTGGG-AGTCT-3′) | 2632-3264 |
| SEH Full F (5′-ACGTCACTAACCACGTGAC-3′) | SEH(1) R (5′-ACAAGATCGCCACAACCAC-3′) | 1-446 |
| SEH(3) F (5′-GGTTCAATTACGGACTGC-3′) | SEH(3) R (5′-TGTTTGATATGGGTCTCTG-3′) | 904-1593 |
| SEH(4) F (5′-TCCGTTCTGCTCACCACAA-3′) | SEH(4) R (5′-TGTGAACTAGAGGGGGATC-3′) | 1376-2163 |
| SEH(5) F (5′-ATGCCACCAAACCTACCACACAGT-3′) | SEH(5) R (5′-AAAGAACCCCCGTCGCCAGTCCCA-3′) | 1975-2558 |
| SEH(6) F (5′-CTCAGTACAAGTCGTTAGC-3′) | SHE Full R (5′-GAAATTCCCGACCC-CCG-3′) | 2480-3292 |
Detection of SEN-V in DNA from human WBCs.
To exclude the possibility that we had amplified the host genome by PCR, we compared the detectabilities of SEN-V DNA from sera and from white blood cells (WBCs). We chose four patients and four controls (two patients each had SEN-V type D- and SEN-V type H-positive sera, and two controls each had SEN-V type D- and SEN-V type H-negative sera) and extracted WBC DNA from all of them. Each pair of samples was randomly chosen from groups of patients whose sera had the same SEN-V infection status. DNA was extracted from 3 ml of whole blood with the SepaGene kit (Sanko Junyaku, Tokyo, Japan) according to the instructions of the manufacturer. The extracted DNA was dissolved in 20 μl of Tris-HCl buffer (10 mM; pH 8.0) containing 1 mM EDTA, heated to 95°C for 15 min, and quickly chilled on ice. The total amount of extracted DNA was subjected to nested PCR. We tried to detect SEN-V DNA in sera and WBCs by PCR, using the same primers and protocol and under the same conditions.
Genetic analysis of SEN-V DNA.
Nucleotide sequencing of detectable SEN-V DNA was performed with an autosequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and the dye termination method as described previously (20). The direct sequencing reactions were carried out with primers specific for genotypes D and H: SED-S3 and SEH-S3, respectively (Table 1). The sequences of SEN-V types D and H were compared to the prototype sequences of SEN-V (SEN-V type D, nucleotides 528 to 677; SEN-V type H, nucleotides 402 to 551) or the reported sequence of TTV (TTV nucleotides 701 to 850 and TTV nucleotides 401 to 550). A phylogenetic tree was constructed by the unweighted pair group method with the arithmetic mean with the computer software package GENETYX-MAC (Software Development Co., Ltd., Tokyo, Japan). Nucleotide sequence homologies among the SEN-V DNAs obtained from patients and controls were calculated and analyzed. In order to identify the full-length nucleotide sequence, fragments of SEN-V types D and H were amplified with the following PCR primer sets: SED Full F and SED(1) R, SED S2 and SED AS2, SED(3) F and SED(3) R, SED(4) F and SED(4) R, SED(5) F and SED(5) R, and SED(6) F and SED Full R and primer sets SEH Full F and SEH(1) R, SEH S2 and SEH AS2, SEH(3) F and SEH(3) R, SEH(4) F and SEH(4) R, SEH(5) F and SEH(5) R, and SEH(6) F and SEH Full R (Table 1). Each primer set was designed to amplify one of six fragments from SEN-V type D (nucleotides 1 to 553, 495 to 953, 868 to 1589, 1439 to 2082, 1990 to 2688, and 2632 to 3264) or one of six fragments from SEN-V type H (nucleotides 1 to 446, 381 to 1163, 904 to 1593, 1376 to 2163, 1975 to 2558, and 2480 to 3292). The full-length nucleotide sequence was identified by direct sequencing with each set of sense and antisense primers. The codon distributions and hydrophobicities of the polyproteins encoded by the open reading frames (ORFs) of SEN-V types D and H were analyzed.
Statistical analysis.
The results are expressed as the means ± standard deviations. Comparisons were performed by Student's t test and the chi-square test. Fisher's exact probability test was used to examine the relationship between HCC and SEN-V. A P value of less than 0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The two full-length nucleotide sequences of SEN-V identified from a CLD patient (type D) and a control (type H) have been submitted to the DDBJ database and given accession numbers AB059532 and AB059353, respectively.
RESULTS
Incidence of SEN-V DNA in HCC and CLD patients.
SEN-V DNA was detected in 25 of 34 (74%) patients with non-B, non-C CLD, 9 of 13 (69%) patients with CLD, and 16 of 21 (76%) patients with HCC. SEN-V DNA was detected in 24 of 32 (75%) controls. The incidence of SEN-V was not significantly different between patients and controls or between patients with HCC and patients with CLD but without HCC.
Clinical features of SEN-V-positive chronic hepatitis and HCC patients.
The clinical features of the patients with HCC and the patients with CLD but without HCC with respect to SEN-V infection status are shown in Table 2. The clinical features included age; sex; serum albumin, total bilirubin, and transaminase levels; the results of the ICG R15 test; platelet count; blood transfusion history; and a history of heavy alcohol intake. There were no statistically significant differences in these clinical features between SEN-V-positive and SEN-V-negative patients. There were no significant differences in tumor markers, such as α-fetoprotein and des-gamma-carboxyprothrombin levels, or in tumor factors (size, tumor-node-metastasis classification) between SEN-V-positive and SEN-V-negative HCC patients (data not shown).
TABLE 2.
Clinical features of patientsa
| Clinical feature | Value for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CLD patients (n = 34)
|
Patients with HCC (n = 21)
|
Patients without HCC (n = 13)
|
|||||||
| SEN-V positive (n = 25) | SEN-V negative (n = 9) | P | SEN-V positive (n = 16) | SEN-V negative (n = 5) | P | SEN-V positive (n = 9) | SEN-V negative (n = 4) | P | |
| Age | 64 ± 12 | 66 ± 9 | 0.74 | 67 ± 10 | 70 ± 8 | 0.69 | 61 ± 15 | 61 ± 8 | 0.77 |
| Sex (no. of M/no. of F) | 22/3 | 7/2 | 0.59 | 15/1 | 3/2 | 0.13 | 7/2 | 4/0 | > 0.99 |
| Serum albumin level (g/dl) | 3.6 ± 0.5 | 3.5 ± 0.4 | 0.59 | 3.3 ± 0.6 | 3.4 ± 0.4 | 0.62 | 3.8 ± 0.4 | 3.7 ± 0.6 | 0.84 |
| Total bilirubin level (mg/dl) | 1.0 ± 0.8 | 1.1 ± 0.7 | 0.91 | 1.3 ± 0.9 | 1.4 ± 0.7 | 0.55 | 0.9 ± 0.6 | 1.0 ± 0.7 | 0.68 |
| AST (IU/liter) | 45 ± 21 | 54 ± 24 | 0.29 | 47 ± 21 | 56 ± 34 | 0.78 | 41 ± 21 | 52 ± 6 | 0.82 |
| ALT (IU/liter) | 41 ± 292 | 38 ± 20 | 0.74 | 42 ± 30 | 39 ± 25 | 0.66 | 40 ± 31 | 36 ± 13 | 0.62 |
| ICG R15 test result (%) | 19 ± 11 | 21 ± 6 | 0.75 | 20 ± 11 | 21 ± 6 | 0.87 | |||
| Platelet count (10,000/μl) | 12.7 ± 5.0 | 11.4 ± 5.4 | 0.53 | 13.0 ± 4.9 | 14.4 ± 2.7 | 0.48 | 11.9 ± 5.5 | 7.6 ± 5.8 | 0.28 |
| BTF historyb | 6/23 (26) | 2/8 (25) | > 0.99 | 4/16 (25) | 0/5 (0) | 0.53 | 2/7 (29) | 2/3 (67) | 0.50 |
| Alcohol use historyb | 14/21 (67) | 4/9 (44) | 0.22 | 12/15 (80) | 2/5 (40) | 0.13 | 2/6 (33) | 1/3 (33) | > 0.99 |
| TTV positiveb | 7/25 (28) | 1/9 (11) | 0.40 | 3/16 (19) | 1/5 (20) | > 0.99 | 4/9 (44) | 0/4 (0) | 0.23 |
Data are expressed as the means ± standard deviations unless indicated otherwise. Abbreviations: M, male; F, female; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BTF, blood transfusion.
The data represent the number of patients positive/number of patients tested (percent).
A background liver biopsy specimen was obtained from 12 of 16 SEN-V-positive HCC patients. Of the 12 SEN-V-positive HCC patients, 5 (42%) had chronic hepatitis and 6 (50%) had cirrhosis. A background liver biopsy specimen was obtained from four of the five SEN-V-negative HCC patients. Of the four SEN-V-negative HCC patients, one (25%) had chronic hepatitis and three (75%) had cirrhosis. The fibrotic stage of background liver disease was not significantly different between SEN-V-positive and SEN-V-negative patients, irrespective of HCC status (P = 0.89).
Both the serum and the WBCs of only one of four SEN-V-positive patients and controls were SEN-V type D positive. However, the WBCs of none of the patients or controls who were SEN-V negative were SEN-V positive.
Genetic analysis of SEN-V DNA.
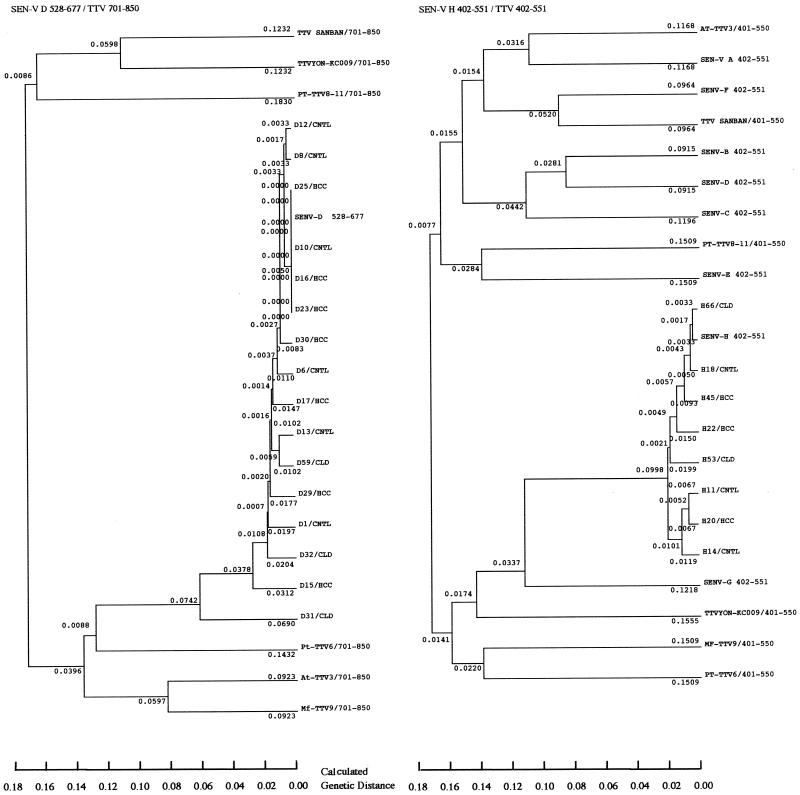
Most of the SEN-V DNA sequences varied between patients, to the extent that we examined them (Fig. 1). D1-13 and H11-18 were SEN-V DNAs obtained from the sera of controls, D15-30 and H20-45 were from the sera of HCC patients, and D31-59 and H53-66 were from the sera of CLD patients without HCC. There were no specific regions where mutations accumulated. All the samples from SEN-V type D- and H-positive patients that were sequenced were classified by genotype according to the genetic analysis shown in the phylogenetic tree. Each genotype-specific primer worked as expected (Fig. 2). All of the clones of SEN-V types D and H obtained were classified as closely related to the TTV family (Fig. 2). The nucleotide sequence homologies among the SEN-V DNAs obtained from patients and controls were calculated. The nucleotide sequence homologies were 80 to 100% (mean ± standard deviation, 93% ± 5%) among SEN-V type D DNAs and 85 to 97% (91% ± 4%) among SEN-V type H DNAs. Two SEN-V type D genomes and one SEN-V type H genome showed comparatively low levels of homology with other sequences, but there were no significant differences among the sequences of SEN-V DNA from controls, HCC patients, and CLD patients without HCC.
FIG. 1.
Multiple-sequence alignments of SEN-V DNAs. The sequences of the samples were compared with the consensus sequence. Only the nucleotides that differed from the consensus sequence are shown. SEN-V type D nucleotides 528 to 677 and SEN-V type H nucleotides 402 to 551 are the consensus sequences. D1, D6, D8, D10, D12, D13, D15, D16, D17, D23, D25, D29, D30, D31, D32, and D59 and H11, H14, H18, H20, H22, H45, H53, and H66 are samples from patients (HCC, CLD) and controls (CNTL).
FIG. 2.
Nucleotides 528 to 677 of SEN-V type D were compared with the sequences of the same regions of SEN-V types A to H and TTV nucleotides 701 to 850. Nucleotides 402 to 551 of SEN-V type H were compared with the sequences of the same regions of SEN-V types A to H and TTV nucleotides 402 to 551. The phylogenetic tree was constructed by the unweighted pair group method with the arithmetic mean by using the computer software package GENETYX-MAC (Software Development Co., Ltd.). The sample numbers are as described in the legend to Fig. 1.
Two full-length nucleotide sequences of SEN-V were identified from a CLD patient and a control. One was type D and the other was type H. These sequences had high degrees of homology with the sequences reported for SEN-V type D and H DNAs (data not shown). The codon distribution and hydrophobicity profile within the ORFs (SEN-V type D, nucleotides 252 to 725; SEN-V type H, nucleotides 256 to 723) of the SEN-V DNAs obtained and reported were analyzed. Differences in codon usage between the SEN-V DNAs obtained and reported were observed for 17 SEN-V type D codons and 18 SEN-V type H codons. The hydrophobicity profiles of amino acids 100 to 110 of the SEN-V D type ORF and amino acids 90 to 100 of the SEN-V type H ORF differed from those reported for SEN-V types D and H.
DISCUSSION
Since the discovery of HCV, it has become obvious that this virus causes the majority of cases of non-A, non-B CLD, including cirrhosis and HCC. However, the causative agent of non-B, non-C CLD remains unknown. Recently, the discovery of SEN-V, a novel DNA virus associated with posttransfusion hepatitis, was reported (19).
The prevalence of SEN-V in our study group of patients with HCC or patients with CLD without HCC is similar to the prevalences reported in previous studies, in which SEN-V DNA was detected in 17 to 92% of patients with liver disease (15, 21-23). However, our study showed a high prevalence of SEN-V in controls, in contrast to the low prevalence (1 to 10%) in subjects without liver disease reported in previous studies (15, 22). The difference in the rate of detection of SEN-V DNA between this and previous reports may be due to differences in the quantity of SEN-V DNA in the sera, differences in the PCR primers used, or differences in the sensitivities of the assay systems used. According to the results of sequence analysis, the specificity of the PCR was high enough. The sensitivity of the PCR could not be determined accurately because the concentration of SEN-V DNA varied among samples. The high rate of detection of SEN-V DNA in sera from healthy controls may indicate a high prevalence of SEN-V. To eliminate the possibility that we were detecting human genome sequences, we performed PCR using WBC DNA as a template.
We also compared the detectabilities of SEN-V DNA from sera and from WBCs. WBCs from none of the SEN-V-negative patients was SEN-V DNA positive. If the SEN-V DNA detected originated in the human genome, it should mainly be detected in WBCs rather than in serum. Furthermore, the sequences of the DNAs that we amplified varied among the patients. The DNA sequences obtained from SEN-V type D- and type H-positive patients were classified by genotype, and those from SEN-V type A to C-positive and SEN-V type E to G-positive patients were classified in the phylogenetic tree. The intragenotype homologies between the sequences obtained from our patients and those reported for SEN-V DNA were at least 87% (SEN-V type D) and 90% (SEN-V type H). These results suggest that the DNA that we detected was viral DNA rather than DNA from the host genome.
We also investigated whether there was a correlation between the presence of SEN-V and the severity of CLD with concomitant HCC. Infection with SEN-V did not have a significant effect on the status of liver disease. The prevalence of SEN-V DNA viremia in our study was similar in patients with HCC (76%) and patients with CLD without HCC (69%). Thus, SEN-V does not seem to contribute to the development of CLD or the development of HCC from CLD.
The prevalence of TTV in this study was lower than that reported previously (18). The difference in the PCR primers used to detect the TTV DNA could explain the lower prevalence of TTV in our study (24).
Genetic analysis of the SEN-V DNA sequences obtained showed no differences among the control, HCC, and CLD without HCC groups. In the analysis of full-length SEN-V type D and H DNAs, we found slight differences in codon distributions and hydrophobicities. In order to estimate whether these differences are significant, we must compare more sequences from strains of the same genotypes.
On the basis of the results of this study, it is unlikely that SEN-V contributes to the incidence of liver disease or the development of HCC in Japan. However, a prospective study of the incidence of HCC in a large population of SEN-V-positive and -negative patients will be required to prove whether SEN-V contributes to the increasing tendency of chronic liver disease to progress to HCC in Japan.
Acknowledgments
We thank H. Kojima for useful discussions.
Financial support was obtained from the Program for the Promotion of Fundamental Studies in the Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan, Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Health Sciences Research Grants for Medical Frontier Strategy Research from the Ministry of Health, Labor, and Welfare.
REFERENCES
- 1.Fukuda, Y., I. Nakano, Y. Katano, T. Kumada, K. Hayashi, S. Nakano, and T. Hayakawa. 1999. TT virus (TTV) is not associated with acute sporadic hepatitis. Infection 27:125-127. [DOI] [PubMed] [Google Scholar]
- 2.Kanda, T., O. Yokosuka, F. Imazeki, M. Tagawa, T. Ehata, H. Saisho, and M. Omata. 1997. GB virus-C RNA in Japanese patients with hepatocellular carcinoma and cirrhosis. J. Hepatol. 27:464-469. [DOI] [PubMed] [Google Scholar]
- 3.Kanda, T., and H. Saisho. 1998. Quantitative analysis of GBV-C RNA in liver and serum by strand-specific reverse transcription-polymerase chain reaction. J. Hepatol. 29:707-714. [DOI] [PubMed] [Google Scholar]
- 4.Kanda, T., O. Yokosuka, T. Ikeuchi, T. Seta, S. Kawai, F. Imazeki, and H. Saisho. 1999. The role of TT virus infection in acute viral hepatitis. Hepatology 29:1905-1908. [DOI] [PubMed] [Google Scholar]
- 5.Kubo, S., S. Nishiguchi, T. Kuroki, K. Hirohashi, H. Tanaka, T. Tsukamoto, T. Shuto, and H. Kinoshita. 1997. Poor association of GBV-C viremia with hepatocellular carcinoma. J. Hepatol. 27:91-95. [DOI] [PubMed] [Google Scholar]
- 6.Leary, T. P., A. S. Muerhoff, J. N. Simons, T. J. Pilot-Matias, J. C. Erker, M. L. Chalmers, et al. 1996. Sequence and genomic organization of GBV-C: a novel member of the Flaviviridae associated with human non-A-E hepatitis. J. Med. Virol. 48:60-67. [DOI] [PubMed] [Google Scholar]
- 7.Lightfoot, M., M. Skelton, M. C. Kew, M. C. Yu, M.-R. Kedda, A. Coppin, and J. Hodkinson. 1997. Does hepatitis GB virus-C infection cause hepatocellular carcinoma in black Africans? Hepatology 26:740-742. [DOI] [PubMed] [Google Scholar]
- 8.Linnen, J., J. Wages, Jr., Z., Y., Zhang-Keck, K. E. Fry, K. Z. Krawczynski, H. Alter, E. Koonin, M. Gallagher, M. Alter, S. Hadziyannis, P. Karayiannis, K. Fung, Y. Nakatsuji, J. W. K. Shih, L. Young, M. Piatak, Jr., C. Hoover, J. Fernandez, S. Chen, J. C. Zou, T. Morris, K. C. Hyams, S. Ismay, Lifson, and G. Hess. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505-508. [DOI] [PubMed] [Google Scholar]
- 9.Love, A., B. Stanzeit, L. Li, E. Olafsdottir, S. Gudmundsson, H. Briem, and A. Widell. 2000. TT virus infections among blood donors in Iceland: prevalence, genotypes, and lack of relationship to serum ALT levels. Transfusion 40:306-309. [DOI] [PubMed] [Google Scholar]
- 10.Nishiguchi, S., M. Enomoto, S. Shiomi, M. Tanaka, K. Fukuda, A. Tamori, T. Tanaka, T. Takeda, S. Seki, Y. Yano, S. Otani, and T. Kuroki. 2000. TT virus infection in patients with chronic liver disease of unknown etiology. Med. Virol. 62:392-398. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa, T., and H. Okamoto. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in post transfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto, H., and T. Nishizawa. 1998. Molecular cloning and characterization of a novel DNA virus (TTV) associated with post transfusion hepatitis of unknown etiology. Hepatol. Res. 10:1-16. [Google Scholar]
- 13.Okuda, K. 1992. Hepatocellular carcinoma: recent progress. Hepatology 15:948-963. [DOI] [PubMed] [Google Scholar]
- 14.Okuda, K., I. Fujimoto, A. Hanai, and Y. Urano. 1987. Changing incidence of hepatocellular carcinoma in Japan. Cancer Res. 47:4967-4972. [PubMed] [Google Scholar]
- 15.Shibata, M., R. Y. Wang, M. Yoshida, J. W. Shin, H. J. Alter, and K. Mitamura. 2001. The presence of a newly identified infectious agent (SEN virus) in patients with liver diseases and in blood donors in Japan. J. Infect. Dis. 184:400-404. [DOI] [PubMed] [Google Scholar]
- 16.Shiratori, Y., S. Shiina, M. Imamura, N. Kato, F. Kanai, T. Okudaira, T. Teratani, G. Togo, N. Toda, M. Ohashi, K. Ogura, Y. Niwa, T. Kawabe, and M. Omata. 1995. Characteristic difference of hepatocellular carcinoma between hepatitis B- and C-viral infection in Japan. Hepatology 22:1027-1033. [DOI] [PubMed] [Google Scholar]
- 17.Simons, J. N., T. P. Leary, G. J. Dawson, T. J. Pilot-Matias, A. S. Muerhoff, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1:564-569. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, K., H. Hoshino, Y. Ohta, N. Yoshida, and S. Mishiro. 1998. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by new sets of PCR primers. Hepatol. Res. 12:233-239. [Google Scholar]
- 19.Tanaka, Y., E. Primi, R. Y. Wang, T. Umemura, A. E. Yeo, M. Mizokami, H. J. Alter, and J. W. Shin. 2001. Genomid and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. J. Infect. Dis. 183:359-367. [DOI] [PubMed] [Google Scholar]
- 20.Togo, G., N. Toda, F. Kanai, N. Kato, Y. Shiratori, K. Kishi, F. Imazeki, M. Makuuchi, and M. Omata. 1996. A transforming growth factor β type II receptor gene mutation common in sporadic cecum cancer with microsatellite instability. Cancer Res. 56:5620-5623. [PubMed] [Google Scholar]
- 21.Umemura, T., H. J. Alter, E. Tanaka, A. E. Yeo, J. W. Shin, K. Orii, A. Matsumoto, K. Yoshizawa, and K. Kiyosawa. 2001. Association between SEN virus infection and hepatitis C in Japan. J. Infect. Dis. 184:1246-1251. [DOI] [PubMed] [Google Scholar]
- 22.Umemura, T., A. E. Yeo, A. Sottini, D. Moratto, Y. Tanaka, R. Y. Wang, J. W. Shin, P. Donahue, D. Primi, and H. J. Alter. 2001. SEN virus infection and its relationship to transfusion-associated hepatitis. Hepatology 33:1303-1311. [DOI] [PubMed] [Google Scholar]
- 23.Willson, L. E., T. Umemura, J. Astemborski, S. C. Ray, H. J. Alter, S. A. Strathdee, D. Vlahov, and D. L. Thomas. 2001. Dynamics of SEN virus infection among injection drug users. J. Infect. Dis. 184:1315-1319. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, H., N. Kato, Y. Shiratori, K. H. Lan, S. K. Ono-Nita, Z. Feng, S. Shiina, and M. Omata. 2000. Poor association of TT virus viremia with hepatocellular carcinoma. Liver 20:247-252. [DOI] [PubMed] [Google Scholar]