Abstract
Various genetic markers have been exploited for fingerprinting the Mycobacterium tuberculosis complex (MTBC) in molecular epidemiological studies, mainly through identifying restriction fragment length polymorphisms (RFLP). In large-scale studies, RFLP typing has practical processing and analysis limitations; therefore, attempts have been made to move towards PCR-based typing techniques. Spoligotyping (spacer oligotyping) and, more recently, variable-number tandem repeat (VNTR) typing have provided PCR-derived typing techniques. This study describes the identification and characterization of novel VNTR loci, consisting of tandem repeats in the size range of 53 to 59 bp in the MTBC, and their assessment as typing tools in 47 Mycobacterium bovis field isolates and nine MTBC strains. Spoligotyping and the previously described set of exact tandem repeats (ETRs) (R. Frothingham and W. A. Meeker-O'Connell, Microbiology 144:1189-1196, 1998) were also applied to the same panel of isolates. The allelic diversity of the individual VNTR loci was calculated, and a comparison of the novel VNTRs was made against the results obtained by spoligotyping and the existing set of ETRs. Eleven unique spoligotypes were discriminated in the panel of 47 M. bovis isolates. Greater resolution was obtained through the combination of the most-discriminating VNTRs from both sets. Considerable discrimination was achieved, with the 47 M. bovis isolates resolved into 14 unique profiles, while all nine MTBC isolates were uniquely differentiated. The novel VNTR markers described increased the discrimination possible in strain typing of M. bovis, with the added benefit of an intuitive digital nomenclature, with the allele copy number of the individual VNTRs providing a profile. VNTR typing was shown to be a valuable technique with great potential for further development and application to epidemiological tracing of tuberculosis transmissions.
Mycobacterium bovis, the causative agent of tuberculosis (TB) in farmed livestock such as cattle and deer, continues to affect detrimentally farming and agricultural economies of many countries, resulting from imposition of trade restrictions and the costs of implementing eradication programs (18). M. bovis is also a recognized zoonotic pathogen that infects many people, particularly in the developing world (35). It also infects a broad range of feral and wildlife animals, such as badgers, deer, goats, and possums; some of these species are reservoirs of infection for farmed livestock (19). An important component in bovine TB eradication programs is the application of epidemiological “traceback.” By developing a better understanding of the source(s) and mode(s) of TB transmission in field outbreaks, more-effective control measures can be implemented (7, 17).
The advent of molecular typing techniques has greatly improved the epidemiological knowledge that can be gained from studying TB outbreaks (1, 17, 20). The three principal M. bovis strain typing techniques described to date have been restriction enzyme analysis (REA) (5), restriction fragment length polymorphism (RFLP) analysis (26), and spoligotyping (11, 13, 22). Standard operating procedures for both REA and RFLP typing of M. bovis have been devised and are generally accepted as providing good levels of differentiation (5, 6, 26, 34). However, there are practical processing and analysis limitations in both REA and RFLP analysis, requiring both skilled personnel and image analysis software. Spoligotyping (spacer oligotyping) is advantageous as it is PCR based and is a more rapid and easier technique to perform and analyze. The main disadvantage of spoligotyping is that all genetic polymorphism is restricted to a single genomic locus, the DR cluster.
The Mycobacterium tuberculosis complex (MTBC) genome sequencing projects at the Sanger Center and The Institute for Genomic Research have released valuable sequence data from the genomes of M. tuberculosis H37Rv, M. bovis AF2122/97, and M. tuberculosis CDC1551, respectively, enabling the identification of polymorphic loci that may be useful for molecular typing (4). Much of the polymorphism occurs within regions of tandemly repeated DNA. Polymorphism at a tandem repeat (TR) locus can occur either as a result of nucleotide sequence changes between individual repeat units or as a result of variation in the number of repeat units, hence creating allelic variants. Variable-number TR (VNTR) typing is based upon repeat number polymorphism within these tandemly arranged repetitive DNA sequences. Many of these TR loci display hypervariablity, enabling their exploitation for strain typing in numerous bacterial species (15, 32).
Previously six VNTR loci, described as exact TR A through F (ETR-A, -B, -C, -D, -E, and -F), were reported and applied to human M. tuberculosis isolates (9). Subsequently, the level of discrimination found between individual M. tuberculosis or M. bovis isolates using the five ETRs (A through E) was not as good as that achieved using either spoligotyping or IS6110-RFLP typing (8, 16). During the course of this investigation, another set of polymorphic repeats, termed mycobacterial interspersed repetitive units (MIRUs), have been described (31). Forty-one MIRU loci were identified within the M. tuberculosis H37Rv genome, of which 12 showed length polymorphisms when applied to a panel of 31 MTBC strains. Additionally, six Queen's University Belfast (QUB) novel VNTRs were identified and applied to 100 M. bovis field isolates (27). Further VNTRs in M. tuberculosis strains H37Rv and CDC1551 were reported from an in silico analysis of their genomes (28). These VNTR markers have not been tested against a panel of isolates; it remains to be seen how many of these are useful.
This study reports 8 TR loci, identified within all three MTBC genomic sequences and their preliminary application to a test panel of 47 M. bovis field isolates for the purpose of strain typing. The discrimination of the novel VNTR loci was compared to the discrimination achieved by spoligotyping and using ETR-A, -B, -C, -D, and -E. In addition, we investigate if it is possible to increase the ability to further resolve the test panel isolates by combining the most discriminative VNTR loci that we report with the ETRs reported previously (9).
MATERIALS AND METHODS
Identification and analysis of TR loci.
TR loci were identified by searching the M. tuberculosis H37RV genomic DNA sequence (accession no. AL 123456) generated at the Sanger Center website (http://www.sanger.ac.uk/Projects/M_tuberculosis) using the software Tandem Repeats Finder, version 2.02 (3). The Tandem Repeats Finder program was downloaded from the web page of the Department of Biomathematical Sciences at Mount Sinai School of Medicine (http://www.c3.biomath.mssm.edu/trf.html). TR loci possessing repeat units ≥50 nucleotides in length, ≥95% nucleotide sequence identity between individual repeat units, and two or more copies of the repeat unit were selected for further analysis. BLAST searches were performed on the M. tuberculosis CDC1551(http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gmt) andthe incomplete M. bovis AF2122/97 (http://www.sanger.ac.uk/Projects/M_bovis) genome sequences to establish the presence and number of repeat units at each TR locus. The VNTR nomenclature convention of identifying of each VNTR locus using the first four digits of its position in the M. tuberculosis H37Rv genome was adopted. BLASTN searches of EMBL and GenBank Bacterial Genome databases for the consensus repeat sequences of the novel VNTRs were performed to determine whether these VNTR loci were unique to the MTBC or related to other repetitive sequences. Nucleotide sequence alignment and cluster analysis were performed on the repeat sequences using DNASIS (version 2.0; Hitachi Software) and BioNumerics (version 2.0; Applied Maths) software. The DNA sequence viewer and annotation tool software Artemis, which is available from the Sanger Center web page, was used to visualize genes flanking the novel VNTRs on the M. tuberculosis H37Rv genome.
Bacterial strains.
A panel of 47 M. bovis field isolates from Northern Ireland (n = 39) and the Republic of Ireland (n = 8) were selected as a test panel. Twenty-seven of the isolates were obtained from bovine hosts, and the remaining 20 isolates were obtained from badgers. Two groups of isolates, A (n = 3) and B (n = 2), were epidemiologically linked, in that they were isolated from animals in the same herd outbreak. A further nine MTBC reference strains were included (see Table 2). These included the M. tuberculosis H37Rv and CDC1551 strains and the M. bovis AF2122/97 strain for which genomic sequence information is available. A further 13 clonally derived M. bovis BCG daughter strains that have been passaged in vitro hundreds of times since 1921 (2) were included (kindly supplied by M. A. Behr [Montreal General Hospital, Montreal, Canada]).
TABLE 2.
VNTR allelic profiles of the M. bovis field isolates and MTBC isolatesa
| Species | Strain(s) | Source(s)b | VNTR allelic profile
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0960 | 2531 | 3232 | 1451 | 1895 | 4156 | 3336 | 1281 | |||
| M. bovis | 030, 033 | ROI | 2 | 4 | 8 | 4 | 2 | 2 | 8 | 2 |
| 005, 036, 044, 045 | NI, ROI | 2 | 4 | 8 | 4 | 3 | 2 | 3 | 2 | |
| 001, 002, 006, 007, 010*, 011*, 013, 014, 015, 016, 017, 025, 026, 029, 032, 034, 035, 037, 040, 041, 047 | NI, ROI | 2 | 4 | 8 | 4 | 4 | 2 | 3 | 2 | |
| 038, 039, 042, 046 | NI | 2 | 4 | 9 | 4 | 2 | 2 | 4 | 2 | |
| 031 | ROI | 2 | 4 | 9 | 4 | 4 | 2 | 10 | 2 | |
| 004, 009, 019, 020, 021♣, 022♣, 023♣, 024, 027, 043 | NI | 2 | 4 | 10 | 4 | 4 | 2 | 3 | 2 | |
| 003, 008 | NI | 2 | 4 | 11 | 3 | 4 | 2 | 4 | 2 | |
| 012 | NI | 2 | 4 | 11 | 4 | 4 | 2 | 6 | 2 | |
| 008 | NI | 2 | 4 | 11 | 4 | 4 | 2 | 8 | 2 | |
| 028 | ROI | 2 | 5 | 12 | 3 | 4 | 2 | 4 | 2 | |
| AF2122/97 | VLA | 2 | 4 | 9 | 4 | 4 | 2 | 3 | 2 | |
| 19210 | ATCC | 2 | 4 | 12 | 4 | 4 | 2 | 10 | 2 | |
| M. bovis BCG | 5692 | NCTC | 2 | 4 | 7 | 4 | 4 | 2 | 12 | 2 |
| P3 | RIVM | 2 | 4 | 6 | 4 | 4 | 1 | 11 | 2 | |
| M. tuberculosis | H37Rv | NCTC | 3 | 6 | 4 | 4 | 4 | 3 | 8 | 2 |
| 14323 | RIVM | 5 | 3 | 22 | 2 | 2 | 4 | 9 | 2 | |
| CDC1551 | CSU | 5 | 5 | 7 | 4 | 2 | 4 | 9 | 2 | |
| M. africanum | 25420 | ATCC | 7 | 4 | 8 | 4 | 4 | 4 | 10 | 2 |
| M. microti | 8710 | NCTC | 5 | 4 | 13 | 4 | 4 | 4 | 21 | 2 |
Isolates that are epidemiologically linked are labeled with like symbols.
Abbreviations: NI, Northern Ireland; ROI, Republic of Ireland; ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures; VLA, Veterinary Laboratories Agency, Weybridge, United Kingdom; RIVM, National Institute of Public Health and the Environment, Bilthoven, The Netherlands; CSU, Colorado State University.
DNA preparation and PCR amplification.
M. bovis isolates were cultured from frozen glycerol or bead stocks on Lowenstein-Jensen slopes containing pyruvate (Media for Mycobacteria, Glamorgan, Wales) for 3 to 4 weeks at 37°C. The MTBC isolates were cultured on Lowenstein-Jensen slopes supplemented with glycerol. A 5-μl sterile loop was used to transfer three to four colonies into 250 μl of sterile distilled water. The suspended colonies were boiled for 5 min in a water bath, and cellular debris was removed from the supernatant by centrifugation at 12,000 rpm in a microcentrifuge (MSE Micro Centaur) for 5 min. PCRs were performed in a final volume of 30 μl. Primer sets that flanked each of the TR loci were designed to enable amplification (Table 1). The primer sets reported previously (9) were used to amplify the ETR-A to -E loci. Each PCR mix contained 3 μl of 10× PCR buffer (Qiagen), 1.5 mM MgCl2, a 0.66 μM concentration of each primer, a 200 μM concentration of each of the four deoxynucleoside triphosphates, 6 μl of 5× Q-solution (Qiagen), and 1 U of HotStarTaq DNA polymerase (Qiagen). Two microliters of template DNA was added to each reaction mixture. Following an initial denaturation at 95°C for 15 min the PCRs were subjected to 30 thermocycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 90 s. Positive and negative control reactions, in which PCR mixes were inoculated with 2 μl of boiled cells from the M. tuberculosis H37RV strain and sterile distilled water, respectively, were performed with each set of reactions. PCR products were electrophoretically separated on 1% NuSieve GTG (FMC, Flowgen) and 1% typing-grade agarose (Life Technologies) gels in 1× Tris-boric acid-EDTA buffer (Invitrogen Life Technologies). A 100-bp DNA Stepladder (Promega) and a 20-bp DNA ladder (FMC, Flowgen) were used as size markers. Allele naming tables for each TR locus were calculated, and allelic variants were named using the copy number of the repeat unit present. Cluster analysis of the allelic profiles generated by VNTR typing was performed using the Bionumerics software package (version 2.0; Applied Maths). The allelic diversities (h) of VNTRs, individually and in combination, were calculated using the following equation: h = 1 − Σxi2 [n/(n − 1)], where n is the number of isolates and xi is the frequency of the ith allele at the locus (25).
TABLE 1.
PCR primer sequence, repeat unit size, and locus position on the M. tuberculosis H37Rv genome and the allelic variants of the orthologous VNTRs in M. tuberculosis CDC1551 and M. bovis AF2122/97 genomes
| VNTR locus | Position on M. tuber- culosis H37Rv | PCR primer sequence (5′→3′) | Description of VNTR locus ina:
|
||
|---|---|---|---|---|---|
| M. tuberculosis H37Rv | M. tuberculosis CDC1551 | M. bovis AF2122/97 | |||
| 0960cb | 960165-960322 | TCGACTTCCAACAGCACCGTCT | (53 bp × 2) + 51 bp | (53 bp × 4) + 51 bp | (53 bp × 2) + 0 bp |
| GCCCACGACCGATAATGGAGC | |||||
| 2531cb | 2531891-2532208 | CGAGTGCTCCGCTCATCTG | (53 bp × 5) + 52 bp | (53 bp × 4) + 52 bp | (53 bp × 4) + 0 bp |
| GGTGTAGCGTCGCTGACCAT | |||||
| QUB 3232 | 3232648-3232864 | CAGACCCGGCGTCATCAAC | (56 bp × 3) + 48 bp | (56 bp × 6) + 49 bp | (56 bp × 4) + 49 bp |
| CCAAGGGCGGCATTGTGTT | |||||
| QUB 1451 | 1451777-1451994 | GGTAGCCGTCGTCGAGAAGC | (57 bp × 3) + 46 bp | (57 bp × 3) + 46 bp | (57 bp × 3) + 46 bp |
| CGCCACCACCGCACTGGC | |||||
| QUB 1895 | 1895351-1895590 | GGTGCACGGCCTCGGCTCC | (57 bp × 4) + 11 bp | (57 bp × 2) + 11 bp | (57 bp × 4) + 0 bp |
| AAGCCCCGCCGCCAATCAA | |||||
| QUB 4156c | 4156796-4156965 | CTGGTCGCTACGCATCGTG | (59 bp × 2) + 51 bp | (59 bp × 3) + 51 bp | (59 bp × 1) + 51 bp |
| TGGTGGTCGACTTGCCGTTGG | |||||
| QUB 3336 | 3336501-3336799 | ATCCCCGCGGTACCCATC | (59 bp × 5) + 0 bp | (59 bp × 6) + 0 bp | (59 bp × 3) + 0 bp |
| GCCAGCGGTGTCGACTATCC | |||||
| QUB 1281c | 1281895-1282015 | GGGGCTGCGGACCTACGGACT | (60 bp × 2) + 2 bp | (60 bp × 2) + 2 bp | (60 bp × 2) + 2 bp |
| CTGGACTCTTGCGGGGACTTCG | |||||
Each VNTR locus was described as (repeat unit size × copy number) + size of the partial repeat. Partial repeats of the whole repeat unit were present towards the 3′ end of some VNTR loci.
VNTR 0960c and VNTR 2531c are also known as MIRU 10c and MIRU 23c, respectively. The suffix “c” indicates that the VNTR locus is in reverse orientation.
Spoligotyping.
Spoligotyping was performed as described previously (13), with the same inoculum used in the VNTR PCR as described earlier. The nomenclature used by the RIVM Institute of Public Health and the Environment, Bilthoven, The Netherlands, for the spoligotypes was adopted.
RESULTS
Identification and characterization of TR loci.
Eight TR loci were identified and selected from the M. tuberculosis H37Rv genome sequence, six of which were novel QUB TRs (Table 1). These TRs were widely distributed about the genome, and all had repeat units ranging in size from 53 to 60 bp. The size and locus positions of the TRs on the M. tuberculosis H37Rv genome and allelic variation of the orthologous loci in the M. tuberculosis CDC1551 and M. bovis AF2122/97 genomic sequences are given in Table 1. Six of the eight TR loci exhibited polymorphism between these genome sequences due to insertion or deletion of whole repeats and hence can be referred to as VNTR loci. A low level of nucleotide sequence degeneracy between individual repeat sequences was also present (data not shown). All eight were located in intergenic positions on the M. tuberculosis H37Rv genome. The genes flanking the novel TR loci encoded products that possessed various putative functions, including kinases, ligases, and transporter and cell division proteins (data not shown). Sequence analysis identified some homology between the novel TR consensus repeat sequences and Mycobacterium leprae genomic sequences. However, no homology was found with other repetitive elements such as polymorphic GC-rich repetitive sequence (24) and the major polymorphic TRs (12).
The nucleotide consensus sequences from each of the eight TR loci were determined and compared by sequence alignment and cluster analysis to previously reported M. tuberculosis TR loci (Fig. 1). Four main subclusters were identified. Cluster A contained QUB VNTRs 4156c, 1451, 3336, 3232, and 1895c in addition to the repeats mss 6, mpp 8, and msx 4, reported previously (28). Cluster B was composed of QUB VNTR 1281 and ETR-B. Cluster C was the largest cluster and contained VNTRs 2531 and 0960c in addition to all the variable MIRUs (30) and the ETR-C to -E repeats (9). VNTRs 2531 and 0960c corresponded to MIRU 23 and MIRU 40, respectively, and as expected clustered together with the other reported MIRU repeats. The final cluster, cluster D, contained ETR-A alone and was the repeat that when compared to the other new and previously reported repeats, exhibited the least nucleotide sequence similarity.
FIG. 1.
Dendrogram of cluster analysis performed on VNTR consensus sequences using global alignment and the unweighted pair-group method using arithmetic averages. Clusters are labeled A to D. ∗, VNTRs described in this study.
Allelic variation using VNTR loci.
Locus-specific PCR primer sets were used to amplify each of the eight TRs from within the genomic DNA of 47 M. bovis field isolates and nine MTBC isolates (Table 2). Ten unique profiles were identified in the M. bovis isolates, and all nine MTBC isolates were differentiated by a unique profile. Seven of the eight TR loci exhibited allelic variation, the exception being TR 1281, at which only a single allele was identified, with a copy number of two.
VNTRs 3232 and 3336 provided the highest level of allelic polymorphism, each identifying five alleles in the M. bovis isolates and with eight and seven different allelic variants, respectively, present within the nine MTBC isolates examined (Table 3). This was reflected in the allelic diversity figures, with VNTR loci 3232 and 3336 achieving h scores of 0.60 and 0.41, respectively (Table 3). However, three of the TR loci (VNTRs 0960, 4156, and 1281c) exhibited no allelic diversity (h = 0) within the M. bovis field isolates, and two others (VNTR 2531 and 1451) exhibited only low levels (h ≤ 0.10). The same panel of isolates was examined using the five VNTR loci ETR-A to -E. When applied to the nine MTBC control isolates alone, the most polymorphic VNTR loci were ETR-A and ETR-D, for which six different alleles were identified (Table 3). However, only the ETR-A and ETR-B VNTR loci exhibited allelic variation when applied to the M. bovis field isolates, displaying lower allelic h scores of 0.40 and 0.37, respectively.
TABLE 3.
Allelic diversity determined by each of the unique VNTRs individually, the VNTR profiles of the best markers combined together and applied to the test panel and spoligotyping.
| Locus(i) or method | No. of alleles
|
Allelic diversity (h)
|
|||
|---|---|---|---|---|---|
| MTBC (n = 9) | M. bovis (n = 47) | M. bovis (n = 47) | M. bovis (n = 42)a | Total panel (n = 56) | |
| 0960cf | 4 | 1 | 0.00 | 0.00 | 0.15 |
| 2531cf | 3 | 2 | 0.02 | 0.02 | 0.09 |
| QUB 3232 | 8 | 5 | 0.60 | 0.58 | 0.65 |
| QUB 1451 | 2 | 2 | 0.10 | 0.11 | 0.12 |
| QUB 1895 | 4 | 3 | 0.34 | 0.38 | 0.35 |
| QUB 4156c | 4 | 1 | 0.00 | 0.00 | 0.18 |
| QUB 3336 | 7 | 5 | 0.41 | 0.44 | 0.55 |
| QUB 1281c | 1 | 1 | 0.00 | 0.00 | 0.00 |
| VNTR 2165 + ETR-A | 6 | 5 | 0.40 | 0.67 | 0.54 |
| VNTR 2461 + ETR-B | 4 | 2 | 0.37 | 0.39 | 0.45 |
| VNTR 0577 + ETR-C | 4 | 1 | 0.00 | 0.00 | 0.15 |
| VNTR 0580 + ETR-D | 6 | 1 | 0.00 | 0.00 | 0.19 |
| VNTR 3192 + ETR-E | 3 | 1 | 0.00 | 0.00 | 0.09 |
| VNTR 3232 + VNTR 3336 | 9 | 9 | 0.65 | 0.65 | 0.75 |
| VNTR 1451 + VNTR 3232 + VNTR 3336 | 9 | 9 | 0.65 | 0.71 | 0.75 |
| VNTR 1895 + VNTR 3232 + VNTR 3336 | 9 | 10 | 0.75 | 0.74 | 0.81 |
| VNTR 1451 + VNTR 1895 + VNTR 3232 + VNTR 3336 | 9 | 10 | 0.75 | 0.74 | 0.81 |
| ETR-A + ETR-B | 8 | 6 | 0.63 | 0.67 | 0.72 |
| ETR-A through -E | 9 | 6 | 0.63 | 0.67 | 0.74 |
| VNTR 1895 + VNTR 3232 + VNTR 3336 + ETR-A + ETR-B | 9 | 14 | 0.87 | 0.90 | 0.90 |
| Spoligotyping | 8 | 11 | 0.66 | 0.71 | 0.74 |
Isolates epidemiologically unrelated to those in previous column.
VNTR 0960c is also known as MIRU 10c, while VNTR 2531c is also known as MIRU 23c.
Stability of the TR loci.
The stability of the eight TR loci was assessed by determining the allelic variation present within 13 clonally derived BCG daughter strains. These BCG strains have been passaged in vitro hundreds of times since 1921, introducing genetic changes that have been observed through previous DNA fingerprinting analysis (2). Six of the TR loci exhibited no allelic variation between the BCG strains. Allelic variation was detected at two of the eight TR loci, VNTR1451 and VNTR 3336. BCG strain Connaught-1948 displayed an allele different from those displayed by the other 12 strains at VNTR locus 1451. The VNTR locus 3336 divided the BCG strains into two distinct groups. BCG strains Brazil-1925, Tokyo-1925, Sweden-1926, Birkhaug-1927, Danish-1931, Prague-1947, and Glaxo-1954 possessed allele 10 at this locus, while the second group, Russia-1924, Tice-1934, Frappier-1937, Phipps-1938, Connaught-1948, and Pasteur-1961, had allele 11.
Spoligotyping.
Eleven unique spoligotypes were identified in the panel of M. bovis isolates (Fig. 2) with a discriminatory capacity, h, of 0.66). The nine MTBC isolates were resolved into eight unique spoligotypes (Table 3).
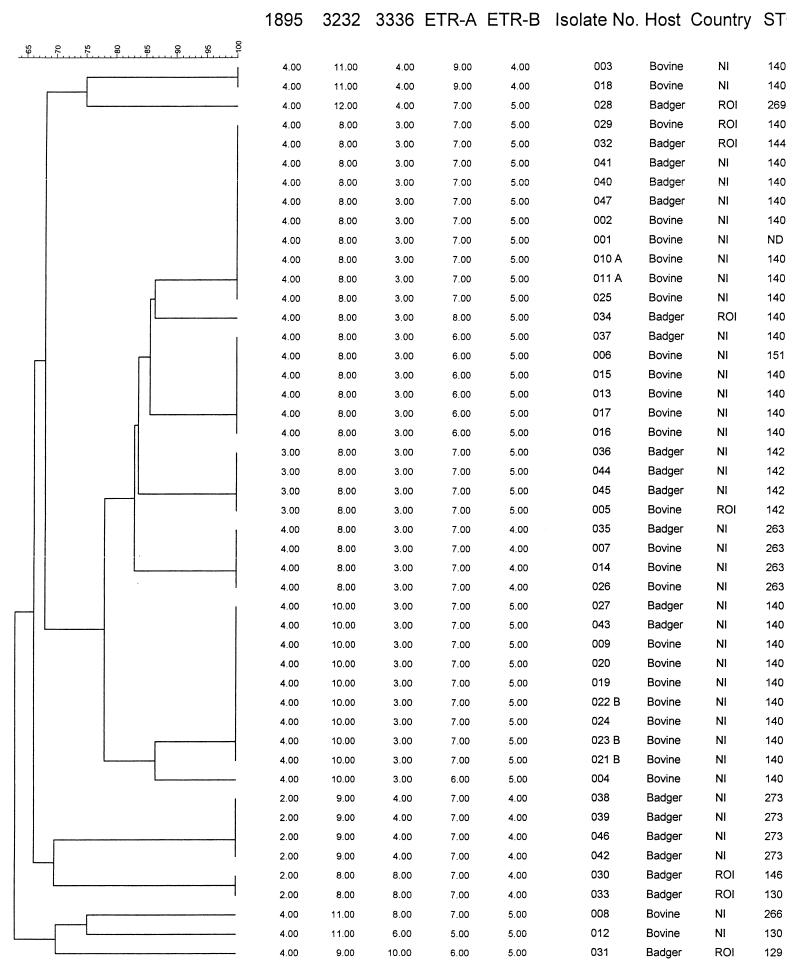
FIG. 2.
Dendrogram constructed by unweighted pair-group method using arithmetic averages using the square root of the categorical similarity coefficient of the 47 M. bovis isolate VNTR profiles with the markers VNTR 1895, 3232, and 3336 and ETR-A and -B. Isolates that are epidemiologically linked are labeled A or B. Abbreviations: NI, Northern Ireland; ROI, Republic of Ireland; ST, spoligotype.
Selection and combination of most-discriminative VNTR loci.
The ability of VNTR typing to distinguish between M. bovis field isolates was improved by combining the four most-discriminating novel QUB loci (VNTRs 3232, 3336, 1895, and 1451) to produce allele profiles (Table 3). The combination of VNTR 3232 and 3336 differentiated the nine MTBC controls into nine unique profiles. When applied to the panel of 47 M. bovis field isolates, VNTRs 3232 and 3336 together differentiated nine allele combinations and reached an h of 0.65. The level of discrimination was further improved by the addition of VNTR 1895, which increased the number of distinct allele profiles to 10 from within the 47 M. bovis field isolates and increased the h to 0.75. Addition of VNTR 1451 to the allele profile had no effect on the level of discrimination between the M. bovis isolates or the allelic diversity. Using the most-variable ETRs (9), ETR-A and -B, the panel of 47 M. bovis field isolates was differentiated into six different allele combinations (Table 3). However, by combining ETR-A and -B with VNTRs 3232, 3336, and 1895, the discrimination was increased, such that the 47 M. bovis field isolates were differentiated into 14 different allele profiles (Fig. 2) and a high (h = 0.87) allelic diversity value was reached. When the two groups of epidemiologically linked M. bovis field isolates were excluded from the analysis, the allelic diversity within the remaining 42 isolates increased (h = 0.90).
Each of the nine MTBC control isolates could be distinguished, having unique allele profiles following application of the five most-discriminating VNTR loci for the 47 M. bovis isolates (VNTRs 3232, 3336, and 1895 and ETR-A and -B). However, eight out of the nine MTBC isolates could be differentiated using either the novel VNTR 3232 alone or the combination of ETR-A and -B. The combination of VNTR 3232 with 3336 differentiated all nine MTBC controls uniquely.
DISCUSSION
In this study, novel polymorphic VNTR loci in the MTBC were identified and characterized. It was demonstrated that these loci were valuable M. bovis strain typing tools, both in their application to field isolates and in the further development of molecular epidemiology. VNTR typing was also shown to be a very promising technique, with excellent potential to be further optimized and developed for application to large-scale epidemiological studies. Molecular variation has previously been exploited to determine evolutionary relationships between many organisms. Genetic markers such as VNTR loci can be used as molecular epidemiological tools to quantitatively examine the similarity and differences between isolates. Epidemiologically linked isolates have been matched through the use of VNTR typing (15, 31). In previous studies, the genomes of MTBC organisms have been found to be genetically homogenous (10, 29). In sequencing 200,000 bp of M. tuberculosis genomic DNA, only four base pair substitutions were observed (14), therefore, VNTR loci are valuable islands of polymorphism. VNTRs have been found in intergenic and nonintergenic regions of genomic DNA and have been found to function as molecular switches in microorganisms, by regulating transcription and possibly translation (21, 33).
In assessing and comparing the novel QUB VNTR loci to other existing VNTR targets, such as ETRs A through E, it was found that one of the newly defined VNTRs, VNTR 3232, displayed a degree of discrimination higher than that found using any of the individual ETRs, while two other novel VNTRs, 3336 and 1895, displayed an equivalent degree of discrimination. It was noted that three of the ETRs, C, D, and E, showed little or no discrimination when applied to M. bovis field isolates. It was also determined that the combination of the most-discriminating VNTR loci from different sets of VNTRs provided the greatest resolution. The three most-discriminating novel VNTR markers described here, combined with two of the most-discriminating ETRs, were shown to resolve the M. bovis isolates better than either of the individual VNTR sets from the different research groups and their discrimination was also greater than that achieved by spoligotyping. VNTR loci making up a set of VNTR markers, described by different research groups, are not necessarily the best combination or set of VNTRs. This work suggests that a better approach is to assess the ability of each individual VNTR to discriminate isolates and to determine the degree of congruence, concordance, and divergence between the different polymorphisms found using particular VNTR markers in combination. Subsequently, the most suitable VNTRs available in the MTBC should be combined together to provide an optimized and standardized set.
Length polymorphisms between M. bovis field isolates were due to insertions and deletions of whole repeat units in the individual VNTR loci. It was also apparent that sequence variations between repeats were also contributing to the polymorphism. The VNTRs were mainly composed of heterogenous repeats, in that nucleotide sequence degeneracy or point mutations were found between individual repeat sequences of the entire VNTR locus (33). The point mutations were randomly distributed throughout the VNTR locus and the individual repeat units. Partial or incomplete repeats were found towards the 3′ end of the VNTR locus, suggesting polarity. Cluster analysis of all the consensus repeat unit sequences of the VNTRs described to date produced four separate groups, one of which contained the MIRUs and ETR-C, -D, and -E; another which contained ETR-A alone; and a third which contained ETR-B and the novel VNTR 1281. The fourth group contained the novel VNTRs that were found to be useful molecular typing tools, VNTRs 1895, 3232, and 3336; these shared a high degree of sequence identity with each other and clustered separately from the MIRUs and ETRs.
Ideally, typing techniques for routine application to large numbers of samples should be rapid and reproducible, require the minimum amount of expensive and specialized equipment or personnel, be cost-effective, and be sufficiently discriminating without destroying concordance. A balance between discrimination and concordance needs to be achieved to allow meaningful epidemiological assessments of relationships to be made between isolates. The novel VNTR loci were found to be exceedingly stable, with six of the loci remaining unchanged in the M. bovis BCG daughter strains, despite hundreds of passages over 30 years (30). The standard operating procedures for both REA and RFLP typing of M. bovis that have previously been devised are generally well accepted as providing good levels of differentiation between M. bovis strain types. However, there are practical processing and analysis limitations in both REA and RFLP analysis when strain typing of large numbers of isolates is involved. They are both time-consuming and labor-intensive techniques that require subculture of M. bovis, a slow growing organism, extraction of genomic DNA, and further manipulation of DNA to obtain analyzable fingerprints. Skilled personnel and image analysis software are required to analyze and interpret the patterns generated by REA and RFLP analysis. The PCR-based techniques, spoligotyping and VNTR typing, are advantageous in that the supernatant of boiled colonies are a sufficient source of template, making it a much more rapid and easier method to analyze. Another advantage of VNTR typing is that the technique is far easier and less laborious to perform than RFLP analysis or spoligotyping. It also has excellent potential to be automated and multiplexed through the use of fluorescently labeled primers (31), with the further possibility of applying the typing technique directly to tissue that has been sequence captured, for simultaneous detection and strain differentiation (13, 23). The VNTR allele profile immediately gives a numerical strain type that can easily be used in a digital nomenclature scheme, which is simple to log into a database and to share and transfer strain type information between laboratories, without any need for coding or an alternative naming system. The highest allelic diversity was found using the five most-discriminating VNTRs (h = 0.90), which was significantly greater than that obtained by spoligotyping (h = 0.66).
Another step towards standardization is in the nomenclature used to describe the TRs. VNTRs can be divided into a number of subsets based on the size of the repeat unit, namely microsatellites or short sequence repeats, minisatellites, and satellite repeat units, which range in size from 1 to ∼15 bp, 10 to 100 bp, and hundreds to thousands of base pairs, respectively. There is a lack of consistency and a level of overlap in the nomenclature used. The novel VNTRs presented in this study are in the minisatellite range; however, we referred to them using the broader term, VNTR. We suggest that a consensus approach to naming or identifying each unique VNTR in the MTBC be considered, rather than each laboratory devising its own acronyms. In this study we adopted a numerical code which consists of the first four digits of the seven-digit number describing the locus position in M. tuberculosis H37Rv, as previously described (28).
Future work will involve automation of the technique to allow greater throughput of isolates, allowing molecular fingerprinting results to be overlaid with epidemiological data available due to the excellent traceability of animal movements in Northern Ireland. It is possible for individual animals, their movements, and the strain type of their M. bovis isolates to be mapped onto the geographical information system, generating greater epidemiological knowledge. Further analysis and optimization of M. bovis VNTR typing by comparing all the individual VNTRs from the previously described sets, with the intention of determining and selecting the best combination, are currently being undertaken. It is also necessary that the results generated through VNTR typing be assessed and compared to those generated by the standardized RFLP typing procedure. The discrimination capacity of the combination of the novel VNTRs described here and the most-discriminating VNTRs described by others (9, 27, 28, 30) should also be assessed in determining the epidemiological relatedness of M. tuberculosis isolates.
Acknowledgments
We thank Julie McCarroll (staff member of the Department of Agriculture and Rural Development for Northern Ireland) and Thomas McCorry (staff member of QUB) for their advice and help. Thanks go to Dick van Soolingen and Petra de Haas (RIVM Institute of Public Health and the Environment) for providing us with M. bovis P3 and M. tuberculosis 14323. Thanks also go to Colorado State University for providing us with M. tuberculosis CDC1551 genomic DNA. Thanks also go to M. A. Behr, Montreal General Hospital, Montreal, Canada, for providing us with the M. bovis BCG daughter strains.
This study was funded by the Department of Environment, Food, and Rural Affairs (DEFRA), London, United Kingdom (grant SE3017).
REFERENCES
- 1.Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, and D. Cousins. 1998. Restriction fragment length polymorphism and spacer oligonucleotide typing: a comparative analysis of fingerprinting strategies for Mycobacterium bovis. Vet. Microbiol. 61:311-324. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., and P. M. Small. 1999. A historical and molecular phylogeny of BCG strains. Vaccine 17:915-922. [DOI] [PubMed] [Google Scholar]
- 3.Benson, G. 1999. Tandem repeats finder: a program to analyse DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McClean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rodgers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Collins, D. M., and G. W. de Lisle. 1985. DNA restriction endonuclease analysis of Mycobacterium bovis and other members of the tuberculosis complex. J. Clin. Microbiol. 21:562-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousins, D. V., R. A. Skuce, R. R. Kazwala, and J. D. A. van Embden. 1998. Towards a standardised approach to DNA fingerprinting of Mycobacterium bovis. Int. J. Tuberc. Lung. Dis. 2:471-478. [PubMed] [Google Scholar]
- 7.Durr, P. A., R. G. Hewinson, and R. S. Clifton-Hadley. 2000. Molecular epidemiology of bovine tuberculosis-I. Mycobacterium bovis genotyping. Rev. Sci. Tech. Off. Int. Epizoot. 19:675-688. [DOI] [PubMed] [Google Scholar]
- 8.Filliol, I., S. Ferdinand, L. Negroni, C. Sola, and N. Rastogi. 2000. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone and in association with spoligotyping. J. Clin. Microbiol. 38:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, S. V., K. Eiglmeier, T. Garnier, R. Brosch, J. Parkhill, B. Barrell, S. T. Cole, and R. G. Hewinson. 2001.Genomics of Mycobacterium bovis. Tuberculosis 81:157-163. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez, M., S. Samper, J.-A. Gavigan, J. F. Garcia Marin, and C. Martin. 1995. Differentiation by molecular typing of Mycobacterium bovis strains causing tuberculosis in cattle and goats. J. Clin. Microbiol. 47:2953-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans, P. W. M., D. van Soolingen, and J. D. A. van Embden. 1992. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J. Bacteriol. 174:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Rapid detection and simultaneous strain differentiation of Mycobacterium tuberculosis for diagnosis and tuberculosis control. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapur, V., L. L. Li, S. Iordanescu, M. R. Hamrick, A. Wanger, B. N. Kreiswirth, and J. M. Musser. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA-polymerase beta-subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 16.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebs, J. R., R. Anderson, T. Clutton-Brock, I. Morrison, D. Young, and C. Donnelly. 1997. Bovine tuberculosis in cattle and badgers. Report by the independent scientific review group. MAFF Publications, London, United Kingdom.
- 18.Neill, S. D., J. M. Pollock, D. B. Bryson, and J. Hanna. 1994. Pathogenesis of Mycobacterium bovis infection in cattle. Vet. Microbiol. 40:41-52. [DOI] [PubMed] [Google Scholar]
- 19.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 20.Perumaalla, V. S., L. G. Adams, J. Payeur, D. Baca, and T. A. Ficht. 1999. Molecular fingerprinting confirms extensive cow-to-cow intra-herd transmission of a single Mycobacterium bovis strain. Vet. Microbiol. 70:269-276. [DOI] [PubMed] [Google Scholar]
- 21.Renders, N., L. Licciardello, C. Ijsseldijk, M. Sijmons, L. van Alphen, H. Verbrugh, and A. van Belkum. 1999. Variable numbers of tandem repeat loci in genetically homogenous Haemophilus influenzae strains alter during persistent colonisation of cystic fibrosis patients. FEMS Microbiol. Lett. 173:95-102. [DOI] [PubMed] [Google Scholar]
- 22.Roring, S., D. Brittain, A. E. Bunschoten, M. S. Hughes, R. A. Skuce, J. D. A. van Embden, and S. D. Neill. 1998. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet. Microbiol. 61:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Roring, S., M. S. Hughes, R. A. Skuce, and S. D. Neill. 2000. Simultaneous detection and strain differentiation of Mycobacterium bovis directly from bovine tissue specimens by spoligotyping. Vet. Microbiol. 74:227-236. [DOI] [PubMed] [Google Scholar]
- 24.Ross, B. C., K. Raios, K. Jackson, and B. Dwyer. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 30:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittan. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skuce, R. A., D. Brittain, M. S. Hughes, L.-A. Beck, and S. D. Neill. 1994. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 32:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skuce, R. A., T. P. McCorry, J. F. Mc Carroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 28.Smittipat, N., and P. Pallittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 29.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 31.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Belkum, A. 1999. The role of short sequence repeats in epidemiologic typing. Curr. Opin. Microbiol. 2:306-311. [DOI] [PubMed] [Google Scholar]
- 34.van Soolingen, D., P. E. W. de Haas, J. Haagsma, T. Eger, P. W. M. Hermans, V. Ritacco, A. Alito, and J. D. A. van Embden. 1994. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J. Clin. Microbiol. 32:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 1993. Report of a W. H. O. meeting on zoonotic tuberculosis (Mycobacterium bovis) with the participation of F. A. O. World Health Organization, Geneva, Switzerland.