Abstract
We report on a strain of Mycobacterium tuberculosis with a deletion in the protein antigen B gene overlapping the probe binding sites for the Abbott Diagnostics LCx M. tuberculosis (LCx-MTB) probe assay. A false-negative result with the LCx-MTB assay delayed a laboratory diagnosis of tuberculosis.
CASE REPORT
The patient was a 47-year-old Canadian-born male who had resided in Australia since the age of 4 years. He presented to a hospital emergency room, where he reported a 2-month history of lethargy, fatigue, night sweats, and a cough productive of sputum. There was no known contact with tuberculosis. The patient consumed more than 50 standard alcoholic drinks weekly and smoked tobacco heavily. Marked weight loss was apparent. Chest radiograph revealed patchy airspace consolidation and cavitation in the left upper lobe. The patient was admitted to the hospital, and three early-morning sputum samples were collected on consecutive days.
Sputum samples were decontaminated by treatment with 4% NaOH for 30 min. Phosphoric acid (1 M) was used to halt digestion and decontamination, and the specimens were concentrated by centrifugation and inoculated onto Lowenstein-Jensen slopes and into BACTEC Middlebrook 12B medium (Becton Dickinson Microbiology Systems, Sparks, Md.). Three concentrated sediments were stained by the Ziehl-Neelson method, which showed 2+ (1 to 10 bacilli in 10 oil immersion fields) acid-fast bacilli (AFB). A diagnosis of pulmonary tuberculosis was considered highly likely, and antituberculosis chemotherapy was commenced.
An LCx-MTB assay (Abbott Diagnostics, Chicago, Ill.) was performed according to the manufacturer's instructions. By relating the LCx-MTB assay results for the specimen to the cutoff value (CO), the presence or absence of Mycobacterium tuberculosis complex DNA was determined. The CO rate was the mean rate of the positive calibrator controls multiplied by 0.30. The S/CO value was the ratio of the sample rate (S) to the CO. A specimen with an S/CO reading equal to or greater than 1 was considered positive, and a specimen with an S/CO reading less than 1 was negative for the presence of M. tuberculosis complex DNA. An S/CO reading of 0.04 was obtained, indicating a negative result. The result was reported by our laboratory as no detection of M. tuberculosis-specific DNA, and the organisms seen in the smear were presumed to be atypical mycobacteria. Antituberculosis therapy was not discontinued despite the negative LCx-MTB result because of a strong clinical suspicion of tuberculosis. The patient was discharged from the hospital and completed standard, supervised therapy consisting of 2 months of treatment with rifampin, isoniazid, ethambutol, pyrazinamide, and pyridoxine, followed by 4 months of rifampin and isoniazid alone. Clinical follow-up revealed a satisfactory recovery with only some residual fibrosis evident on chest radiographs.
The demand for rapid and reliable methods for the direct detection and differentiation of M. tuberculosis from atypical mycobacteria has led to direct nucleic acid amplification tests being widely applied as routine diagnostic tools in clinical laboratories. The development of PCR and other nucleic acid amplification has led to the introduction of in-house and commercial assays for the detection of M. tuberculosis DNA directly from processed clinical samples. These methods demonstrate high sensitivity (95 to 99%) in detecting M. tuberculosis-specific nucleic acids in processed respiratory specimens with positive AFB smears (2, 5, 6). We report a case of AFB smear-positive pulmonary tuberculosis which yielded a false-negative result in an LCx-MTB assay due to a deletion of the target region in the 38-kDa protein antigen B gene of the strain.
The strain was isolated from BACTEC 12B medium after 12 days of incubation, and direct microscopy revealed AFB showing good serpentine cording. The organism was confirmed as M. tuberculosis complex by M. tuberculosis-specific probe (AccuProbe; GenProbe, San Diego, Calif.) and by MPB70 PCR (10). Species-level identification tests identified the organism as M. tuberculosis. The organism accumulated niacin, produced pyrazinamidase, reduced nitrate, and was susceptible to 10 μg of thiophene-2-carbooxylic acid hydrazide/ml (8, 9). Susceptibility testing was performed by the BACTEC 460 radiometric assay proportion method, according to the BACTEC 460 product and procedure manual (revision E), and the strain was susceptible to rifampin, isoniazid, ethambutol, and streptomycin. A stored aliquot of the sediment was retested by the LCx-MTB assay in parallel with an aliquot of sediment spiked with a control strain of M. tuberculosis (H37Rv). A negative LCx-MTB result was obtained on retesting the sediment, and the spiked sample showed no evidence of inhibition. A sample of the culture was tested by the LCx-MTB assay, and a negative result was obtained. The test was repeated with the culture spiked with an aliquot of H37Rv to exclude the possibility that the culture was inhibitory to the assay, and a positive result was observed.
As the coordinating laboratory for the 2001 Australian Society of Microbiology Mycobacteriology Quality Assurance Programme, we distributed this strain to several laboratories in Australia and New Zealand as a spiked sputum sediment for direct nucleic acid testing for M. tuberculosis. Laboratories using in-house IS6110 PCR methods or the Roche Amplicor system detected M. tuberculosis-specific DNA in the sample. All laboratories using the LCx-MTB assay were unable to detect the M. tuberculosis-specific DNA. We postulated there was a mutation in one or more of the probe binding sites and proceeded to investigate the reason for the false-negative result.
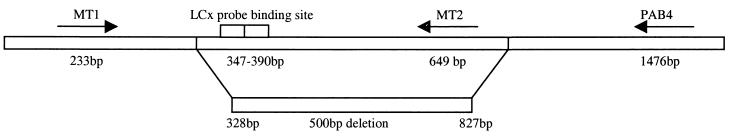
The target nucleic acid sequence for the LCx-MTB assay is found within the single-copy chromosomal gene for protein antigen B (1). The LCx-MTB assay uses four oligonucleotide probes labeled with either a capture or detection hapten to amplify a 44-bp fragment of the protein antigen B gene (bases 347 to 390) (4). The previously described primers MT1 and MT2 (7) were used to amplify a 419-bp product overlapping the 44-bp target of the LCx-MTB assay. PCR was performed as previously described (7) and yielded a 419-bp product with H37Rv and no product with the LCx-MTB-negative strain. An additional primer, pab4, was designed by using Primer Express version 1.5 (Applied Biosystems, Foster City, Calif.) outside the 419-bp region of the protein antigen B gene: 5′-GAGCCTGATCGCACCCATC-3′. PCR was performed with a combination of the primers MT1 and pab4, and a product of 744 bp in the patient strain was detected when the expected product size was 1,244 bp.
The double-stranded 744-bp product was purified with a NucleoSpin extract column (Macherey-Nagel, Duren, Germany), and the sequencing reaction was performed with the ABI Big Dye terminator cycle sequencing ready reaction kit (Applied Biosystems) using 3.2 pmol of sequencing primers MT1 and pab4. The sequencing product was purified with Centricep columns (Princeton Separations, Adelphia, N.J.), and the reaction mixture was loaded onto an ABI Prism 310 genetic analyzer in accordance with the manufacturer's instructions (Applied Biosystems). The results of the sequence reaction were analyzed with Sequencher version 3.0 (Gene Codes Corporation, Ann Arbor, Mich.). Analysis was performed by comparing sequences obtained against GenBank accession no. M30046. A 500-bp deletion in the protein antigen B gene corresponding to bases 328 to 827 was detected. Figure 1 indicates the locations of the primers used, the probe binding sites for the LCx probes, and the region of deletion detected in the protein antigen B gene (GenBank accession no. M30046) of this strain.
FIG. 1.
Schematic diagram showing the LCx-MTB assay probe binding sites, primer binding sites (MT1, MT2, and pab4), and the region of deletion in the protein antigen B gene of a strain of M. tuberculosis.
M. tuberculosis strain typing was performed by the method of Frothingham and Meeker-O'Connell (3), targeting five exact tandem repeat loci (ETR), ETR A to ETR E, to obtain a numerical profile for the strain based on variable numbers of tandem DNA repeats (VNTR) at each locus. A VNTR profile of 42453 was obtained. We examined our database of VNTR profiles (1999 to 2000) to determine if there were other M. tuberculosis strains with the same VNTR profile which could harbor the same deletion. We found only one epidemiologically unrelated strain with the same VNTR profile. M. tuberculosis DNA could be detected in this strain with the LCx-MTB assay, indicating that the deletion in the protein antigen B gene was not present.
The LCx-MTB assay has been in use in our laboratory and others for several years and has proved to be a highly reliable test for detecting M. tuberculosis-specific DNA in smear-positive respiratory specimens (5). All nucleic acid testing methods have the potential to give false-negative results due to mutations which may arise at primer or probe binding sites. These nucleic acid amplification results therefore need to be interpreted in relation to other patient or clinical information. Our patient in this case was unaffected by the delayed laboratory diagnosis of tuberculosis, as treatment for tuberculosis proceeded despite a negative LCx-MTB assay.
REFERENCES
- 1.Andersen, A. B., and E. B. Hansen. 1989. Structure and mapping of antigenic domains of protein gene b, a 38,000-molecular-weight protein of Mycobacterium tuberculosis. Infect. Immun. 57:2481-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Amato, R. F., A. A. Wallman, L. H. Hochstein, P. M. Colaninno, M. Scardamaglia, E. Ardila, M. Ghouri, K. Kim, R. C. Patel, and A. Miller. 1995. Rapid diagnosis of pulmonary tuberculosis by using Roche AMPLICOR Mycobacterium tuberculosis PCR test. J. Clin. Microbiol. 33:1832-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 4.Lindbrathen, A., P. Gaustad, B. Hovig, and T. Tonjum. 1997. Direct detection of Mycobacterium tuberculosis complex in clinical samples from patients in Norway by ligase chain reaction. J. Clin. Microbiol. 35:3248-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lumb, R., K. Davies, D. Dawson, R. Gibb, T. Gottlieb, C. Kershaw, K. Kociuba, G. Nimmo, N. Sangster, M. Worthington, and I. Bastian. 1999. Multicentre evaluation of the Abbott LCx Mycobacterium tuberculosis ligase chain reaction assay. J. Clin. Microbiol. 37:3102-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noordhoek, G., A. Kolk, G. Bjune, D. Catty, J. Dale, P. Fine, P. Godfrey-Faussett, S. Cho, T. Shinnick, S. Svenson, S. Wilson, and J. van Embden. 1994. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison of seven laboratories. J. Clin. Microbiol. 32:277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjobring, U., M. Mecklenburg, A. B. Andersen, and H. Miorner. 1990. Polymerase chain reaction for the detection of Mycobacterium tuberculosis. J. Clin. Microbiol. 28:2200-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wayne, L. G. 1974. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 109:147-151. [DOI] [PubMed] [Google Scholar]
- 9.Wayne, L. G., H. W. B. Engel, C. Grassi, W. Gross, J. Hawkins, P. A. Jenkins, W. Kappler, H. H. Kleeberg, I. Krasnow, E. E. Nel, S. R. Pattyn, P. A. Richards, S. Showalter, M. Slosarek, I. Szabo, I. Tarnok, M. Tsukamura, B. Vergmann, and E. Wolinsky. 1976. Highly reproducible techniques for use in systematic bacteriology in the genus Mycobacterium: tests for niacin and catalase and for resistance to isoniazid, thiophene 2-carboxylic acid hydrazide, hydroxylamine, and p-nitrobenzoate. Int. J. Syst. Bacteriol. 26:311-318. [Google Scholar]
- 10.Wilton, S., and D. Cousins. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1:269-273. [DOI] [PubMed] [Google Scholar]