Abstract
Although the 5′ untranslated region (5′ UTR) is the most conserved region of the hepatitis C virus (HCV) genome, it has been suggested that interrogation of this region is sufficient for determination of the HCV genotype. We compared two methods of determination of the HCV genotype: (i) direct sequencing of the DNA of the NS-5b region and (ii) reverse line probe assay (LiPA; INNO-LiPA HCV II; Innogenetics N.V.) of the 5′ UTR. There was 100% concordance between the two methods for genotype but only 80% concordance for subtype. A significant percentage of genotype 1a isolates were misclassified by LiPA as genotype 1b. Sequence analysis revealed that the only consistent difference in the 5′ UTR for these genotype 1a isolates misclassified as genotype 1b was a single nucleotide (A/G) at position −99 of the HCV genome. All isolates with discordant results analyzed had a G at this position, consistent with LiPA determination of these samples as subtype 1b. However, sequence analysis of 222 nucleotides in the NS-5b region clearly identified all of these isolates as subtype 1a. Population distribution data from the University of Pittsburgh Medical Center of over 200 samples analyzed by sequencing of the NS-5b region and over 1,000 samples analyzed by LiPA also indicated that INNO-LiPA HCV II cannot accurately differentiate HCV genotype 1a isolates from HCV genotype 1b isolates. We provide evidence that the A/G at position −99 represents a sequence polymorphism in the HCV genome that cannot differentiate subtype 1a from subtype 1b isolates. In conclusion, the 5′ UTR is not heterogeneous enough for use in determination of the HCV subtype and cannot be used for differentiation of HCV genotypes 1a and 1b.
Hepatitis C virus (HCV) is an enveloped positive single-stranded RNA virus which is the major cause of chronic hepatitis worldwide (2, 12). Chronic hepatitis resulting from HCV infection may lead to severe sequelae, including hepatic cirrhosis and hepatocellular carcinoma (3). HCV demonstrates a high degree of sequence variation throughout its genome and exists in vivo as a group of heterogeneous but closely related quasispecies (8, 20). However, the levels of heterogeneity differ considerably among the various regions of the virus, ranging from as little as 10% in the 5′ untranslated region (5′ UTR) to 50% or more within the E1 region (8, 10, 32). Based on the analysis of variable coding regions in the viral genome, distinct genotypes as well as subtypes have been defined (10, 16, 34). The distribution of HCV genotypes differs geographically, with subtype 1a being most common in the United States, subtype 1b being most common in Europe and Japan, and other genotypes being prevalent in other parts of the world (for a review, see reference 41). Several studies have shown that HCV genotype influences the response to therapy with alpha interferon alone or in combination with ribavirin (1, 14, 23, 43). In general, HCV type 2 and 3 isolates have higher rates of response to therapy than type 1 isolates. In addition, some studies suggest that genotype alone may predict disease severity; e.g., patients infected with HCV genotype 1a or 1b may exhibit more severe liver disease (4, 6, 18, 27-31, 42-44). These findings indicate an important role of genotype identification for prediction of the outcome of HCV infection and the selection of patients for treatment protocols.
Identification of the HCV genotype by interrogation of several different regions of the HCV genome has been reported. A wealth of phylogenetic information has been derived from sequence analysis of the NS-5 gene of HCV (10, 16, 34). We have determined HCV genotypes using direct sequencing of the DNA of the NS-5b region for several years and have devised an algorithm of sequence analysis for genotype determination on the basis of the sequence of this region. Although sequence analysis is considered the “gold standard” for HCV genotype determination, it is expensive, time-consuming, and inconvenient for routine use. A commercial assay is available which allows rapid analysis for the HCV genotype via a reverse hybridization line probe assay (LiPA) of the 5′ UTR of HCV (INNO-LiPA HCV II; Innogenetics N.V., Zwijnaarde, Belgium). The 5′ UTR is the most highly conserved region among HCV strains (9, 19, 26, 35). This makes this region theoretically superior for sensitivity but limits its usefulness for determining differences between various subtypes. However, phylogenetic analysis of sequence information obtained from the 5′ UTR has correlated fairly well with phylogenetic analysis of sequence information obtained from other regions of the viral genome, including the core, NS-3, NS-4, and NS-5 regions (10, 15, 36, 37). In this study, we compared the HCV genotypes determined by direct sequencing of the DNA of the NS-5b region and LiPA analysis of the 5′ UTR for 63 samples and report our experience with genotyping of over 1,000 samples analyzed by one or both methods over 5 years.
MATERIALS AND METHODS
Samples.
Fresh plasma or liver biopsy specimens sent to the Laboratory of Molecular Diagnostics at the University of Pittsburgh Medical Center for routine HCV genotype analysis over the past 5 years were analyzed. A total of 247 samples were analyzed by NS-5b sequence analysis from 1996 to 1998; a total of 1,035 samples were analyzed by LiPA from 1998 to 2001. A total of 63 samples that were representative of the genotypes identified in our laboratory were initially selected for comparison of genotyping by the two different methods. Sequencing analysis of both the NS-5b region and the 5′ UTR was performed for a number of samples for which LiPA gave discrepant or indeterminate results. Analysis of these samples was done in compliance with federal and institutional review board policies.
cDNA preparation.
RNA was extracted from 230 μl of plasma or 3-mm3 liver biopsy samples with the TRIzol reagent (GIBCO-BRL [now Invitrogen], Carlsbad, Calif.), precipitated with isopropanol, and resuspended in 20 μl of diethyl pyrocarbonate-treated water. cDNA was prepared from 3 μl of plasma RNA or 500 ng of liver RNA. The RNA was heated for 5 min at 70°C and quick chilled on ice; and then 7 μl of a pre-master mixture containing 22 pmol of random hexamers (GIBCO-BRL), 50 mM Tris-HCl (pH 8.3), 7.5 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.2 mM (each) deoxynucleoside triphosphate (Pharmacia, Peapack, N.J.), and 20 U of RNase inhibitor (Promega, Madison, Wis.) was added. A total of 100 U of Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL) was added, and the mixture was incubated at 37°C for 60 min.
LiPA.
RNA purification and cDNA preparation were performed as described above. Analysis of the 5′ UTR was then carried out with the INNO-LiPA HCV II kit (Innogenetics N.V.) according to the instructions of the manufacturer. Briefly, biotinylated primers are used to amplify the 5′ UTR region of HCV by PCR. The biotin-labeled PCR products are then reverse hybridized to type-specific probes attached to nitrocellulose strips. Development results in a purple or brown precipitate as a positive line on the strip. The HCV type and subtype were deduced on the basis of the patterns of hybridizing bands by using the INNO-LiPA HCV II interpretation chart.
PCR of the NS-5b and 5′ UTR regions.
PCR amplification was performed with the primers described below in a model 9600 thermocycler (Perkin-Elmer) with 10 μl of cDNA in a total reaction volume of 50 μl containing each primer at 10 pmol, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, and 1.0 U of Taq DNA polymerase (Perkin-Elmer). After an initial denaturation step at 95°C for 1 min, 40 cycles of PCR at 95°C for 20 s, 56°C for 30 s, and 72°C for 1 min were carried out, with a final extension of 72°C for 10 min. Nested PCR of the NS-5b region was performed with 1 μl of the first-round amplification product if the amplification product was not sufficient for sequence analysis. Twenty-five cycles of the nested PCR were carried out by using the same conditions described above, except that an annealing temperature of 60°C was used. First-round amplification of the NS-5b region was done with a sense primer specific to bases 7904 to 7922 (5′-TGG GGT TCT CGT ATG ATA CCC-3′) and an antisense primer specific to bases 8295 to 8275 (5′-CCT GGT CAT AGC CTC CGT GAA-3′). Nested PCR primers for detection of the NS-5b region were a sense primer specific to bases 7916 to 7935 (5′-GAT ACC CGC TGC TTT GAC TC-3′) and an antisense primer specific to bases 8284 to 8266 (5′-CCT CCG TGA AGG CTC TCA G-3′). PCR primers for detection of the 5′ UTR were a sense primer specific to bases −333 to −317 (5′-CCG ATT GGG GGC GAC AC 3′) and an antisense primer specific to bases 05 to −14 (5′-CTC ATG GTG CAC GGT CTA C-3′). The nucleotide numbering for the primers is according to the prototype HCV type 1 sequence (13). All primers were manufactured by GIBCO-BRL.
Direct sequencing of PCR products.
PCR products were purified with MicroCon-100 filters and were sequenced with standard or Big Dye terminator kits (Perkin-Elmer) on an automated ABI373 or ABI377XL instrument. The sequencing primer for the NS-5b region was a sense primer specific to bases 7916 to 7935 (5′-GAT ACC CGC TGC TTT GAC TC-3′). Sequencing of the 5′ UTR was performed in both directions for 6 samples that gave discrepant results by the two methods and for 10 samples that gave indeterminate results by LiPA by using a sense primer specific to bases −333 to −317 (5′-CCG ATT GGG GGC GAC AC-3′) and an antisense primer specific to bases 05 to −14 (5′-CTC ATG GTG CAC GGT CTA C-3′).
Sequence analysis of the NS-5b region.
We have devised an algorithm for determination of HCV genotype based on sequence analysis of the NS-5b region (Z. Chen and J. A. Kant, unpublished data). Briefly, by using consensus nucleotide sequences from a 222-bp region within the HCV NS-5b region (34), three or four key nucleotides that are specifically associated with major types of HCV were first selected. These nucleotides are shared by each subtype within major types. Another 5 to 11 key nucleotides specifically associated with subtypes within major HCV types were then chosen. Determination of the type and subtype of HCV in each patient sample was made on the basis of the presence of these key nucleotides that had been identified. In addition, the alignments of the sequenced 222-bp region with the NS-5b region was determined for each patient sample by using consensus HCV subtype sequences (34), and the total number of mismatches per 222 bp was determined for each subtype within the assigned type. Typically, we find fewer than 15 mismatches per 222 bp for samples of the same HCV subtype (>93% similarity) and greater than 30 mismatches per 222 bp for samples of different HCV subtypes (<86% similarity). This is consistent with proposed similarities for classification of HCV genotype on the basis of the sequence of the NS-5 gene (33). For all samples analyzed, determination of genotype based on this algorithm correlated with the results obtained by sequence analysis with the BLAST program.
Restriction enzyme digestion.
Restriction enzyme analysis of the 5′ UTR was performed for a subset of samples that were genotype 1 by LiPA but whose subtypes could not be determined. A total of 20 μl of purified reverse transcription-PCR (RT-PCR) products from the 5′ UTR were digested with Alw26I at 37°C for 3 h and analyzed on a 2% agarose gel stained with ethidium bromide.
RESULTS
Comparison of HCV genotype by NS-5b sequence analysis and LiPA.
A total of 63 samples were analyzed by both NS-5b sequence analysis and LiPA in a comparative analysis (Table 1). The genotypes of the samples selected for the comparative analysis were representative of the genotypes identified in our laboratory by NS-5b sequencing in order to evaluate the performance of the INNO-LiPA HCV II kit. There was 100% concordance between the two methods for type and 80% concordance for subtype. LiPA failed to assign a subtype to four (6%) samples. Of the samples that were assigned a subtype by both methods, there were discrepant subtyping results for 9 of 63 (14%) samples. One sample was called subtype 2b by NS-5b sequence analysis and subtype 2a/2c by LiPA. LiPA cannot discriminate between subtypes 2a and 2c. The remainder of the samples with discrepant results (13% of the total) were called subtype 1b by LiPA and subtype 1a by NS-5b sequence analysis. Of the 37 samples identified as subtype 1a by NS-5b sequence analysis, 22% were classified as subtype 1b by LiPA.
TABLE 1.
Comparison of HCV genotype by NS-5b sequence analysis versus LiPAa
| No. (%) of isolates | Genotype by:
|
|
|---|---|---|
| NS-5b sequence analysis | LiPA | |
| 28 (44) | 1a | 1a |
| 7 (11) | 1b | 1b |
| 8 (13) | 1a | 1b |
| 1 (1.6) | 1a | 1 |
| 1 (1.6) | 2a | 2a/2c |
| 7 (11) | 2b | 2b |
| 1 (1.6) | 2b | 2a/2c |
| 9 (14) | 3a | 3a |
| 1 (1.6) | 4a | 4 |
| 63 | ||
The genotyping results for 63 samples that were selected for analysis by both methods in a comparative analysis are shown. Isolates with discrepant results are in boldface.
NS-5b sequence analysis of samples with discrepant results.
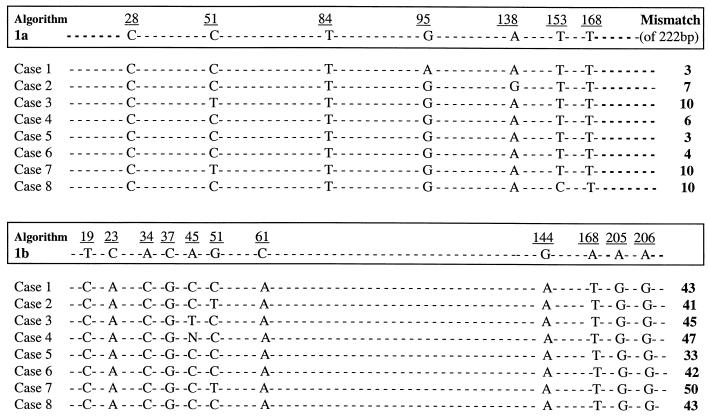
Figure 1 shows the results of NS-5b sequence analysis for the eight samples with discrepant results that were subtyped as HCV subtype 1b by LiPA and subtype 1a by NS-5b sequence analysis. All of the samples with discrepant results were sequenced at least twice in order to confirm their classification as type 1a. The eight samples each demonstrated six to seven of the seven nucleotides identified as key nucleotides for genotype 1a. In contrast, none of these samples had any of the 11 nucleotides identified as key for genotype 1b. In addition, the alignments of the sequences with consensus sequences for subtypes 1a and 1b, as described by Simmonds et al. (34), were compared. The total number of mismatches was significantly higher with subtype 1b consensus sequences than with subtype 1a consensus sequences for all eight isolates analyzed. This corresponds to 95.5 to 98.6% nucleotide identity with the subtype 1a consensus sequence and 77 to 85% nucleotide identity with the subtype 1b consensus sequence, thus confirming the identification of all eight of these samples as HCV genotype 1a.
FIG. 1.
NS-5b sequence analysis of samples with discrepant results. The algorithm used to define HCV genotype by NS-5b sequence analysis is described in Materials and Methods. The key nucleotides associated with subtypes 1a and 1b are numbered and boxed. Sequence results are shown only for these key nucleotides. Nucleotide 01 in the algorithm corresponds to nucleotide 7975 in the HCV type 1 prototype sequence (13). The rightmost column shows the total number of mismatches of 222 bp analyzed compared to consensus subtype 1a or subtype 1b sequences (34).
5′ UTR sequence analysis of samples with discrepant results.
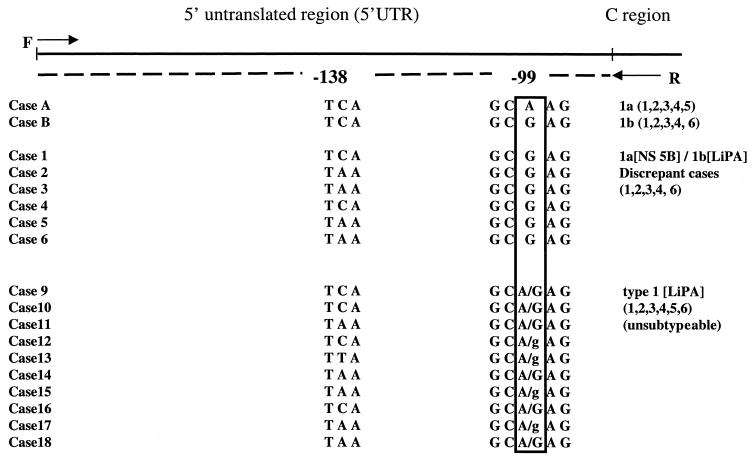
In order to investigate the discrepancy further, we sequenced the DNA of the 5′ UTR region (the region interrogated by LiPA) for six of the eight samples with discrepant results for subtype 1a and 1b and for control subtype 1a and subtype 1b samples (Fig. 2). The only consistent difference in the 5′ UTR for these samples was a single nucleotide (A/G) at position −99 of the HCV genome. Determination of a sample as subtype 1a or 1b by LiPA is on the basis of this single A/G difference at position −99, with LiPA probe 5 corresponding to an A residue at nucleotide −99 and LiPA probe 6 corresponding to a G residue at this position (38). All six samples with discordant results analyzed had a G residue at this position, consistent with identification of these samples as subtype 1b by LiPA analysis of the 5′ UTR. However, all six samples were clearly subtype 1a by NS-5b sequence analysis (Fig. 1).
FIG. 2.
DNA sequencing analysis of the 5′ UTR for control subtype 1a and subtype 1b samples (cases A and B), for six of the samples from Fig. 1 with discrepant results for subtypes 1a and 1b (cases 1 to 6), and for 10 samples that LiPA classified as type 1 but that were unsubtypeable by LiPA (cases 9 to 18). All of the samples had identical 5′ UTR sequences with the exception of the nucleotide regions at positions −99 and −138. The banding pattern obtained by LiPA is shown in parentheses at the right. C region, core region; F, forward sequencing primer; R, reverse sequencing primer; A/G, both an A-residue peak and a G-residue peak were observed at an approximately equal peak ratio by sequence analysis; A/g, a higher peak was observed for the A residue than for the G residue by sequence analysis.
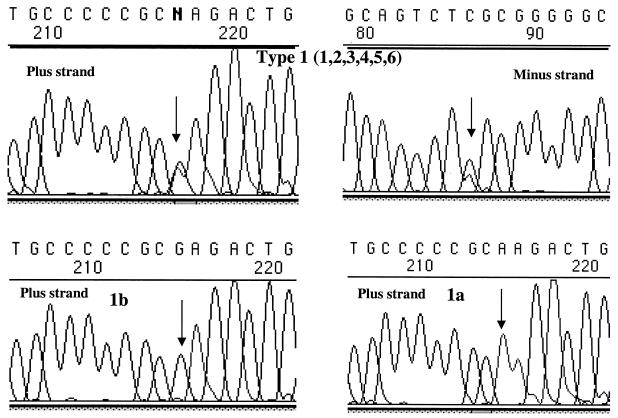
One of the samples with discrepant results listed in Table 1 was subtype 1a by NS-5b sequence analysis but genotype type 1 by LiPA and could not be subtyped by LiPA. Subsequent analysis of over 1,000 samples by LiPA in our laboratory indicated that a significant proportion (7.7%) of HCV genotype 1 samples cannot be subtyped by LiPA (see Fig. 5). Many of these have LiPA band pattern 1, 2, 3, 4, 5, 6. In order to investigate these further, DNA sequencing analysis was performed for 10 samples that were classified as type 1 by LiPA but that could not be subtyped by LiPA. The genotypes of all 10 of these were unambiguously subtype 1a by sequencing of the NS-5b gene (data not shown). However, sequence analysis of the 5′ UTR of these 10 samples with LiPA band pattern 1, 2, 3, 4, 5, 6 demonstrated a heterogeneous pattern, with both A and G nucleotides identified at position −99 (Fig. 2 and 3). In some samples, almost equal proportions of both nucleotides were present (a heterozygous pattern); in other samples, the peak for one nucleotide was higher than that for the other.
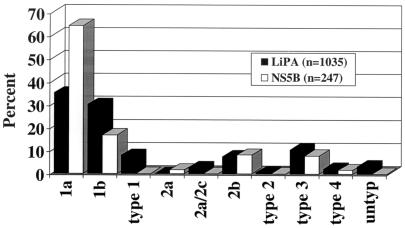
FIG. 5.
Comparison of the distributions of HCV genotypes obtained by LiPA versus those obtained by NS-5b sequence analysis.
FIG. 3.
Representative sequence analysis for HCV subtype 1a and 1b controls and for a type 1, unsubtypeable isolate with LiPA banding pattern 1, 2, 3, 4, 5, 6. Position −99 is indicated by the arrows. Samples found to be subtype 1a by LiPA have an A residue at this position; samples found to be subtype 1b have a G residue. The heterogeneous pattern observed at this nucleotide position for cases 9 to 18 from Fig. 2 is demonstrated. Forward (plus-strand) or reverse (minus-strand) sequences are indicated.
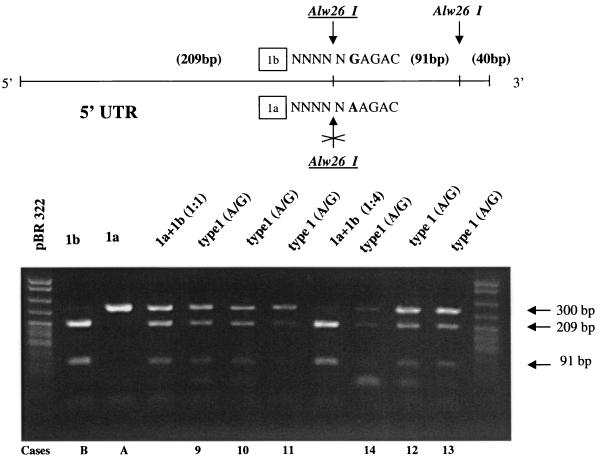
Digestion of RT-PCR products of type 1 isolates with Alw26I.
The 5′ UTR RT-PCR products of six of the type 1 samples whose sequences are presented in Fig. 2 as well as those of subtype 1a and subtype 1b controls were subjected to restriction enzyme digestion with Alw26I in order to differentiate between a G residue at position −99 (subtype 1b pattern) and an A residue at this position (subtype 1a pattern) (Fig. 4). Restriction enzyme digestion of all six type 1 samples with LiPA band pattern 1, 2, 3, 4, 5, 6 demonstrated a heterogeneous pattern. These results confirmed the results of sequencing analysis of the 5′ UTR that samples with LiPA banding pattern 1, 2, 3, 4, 5, 6 carry both an A residue and a G residue at position −99 (Fig. 2 and 3) and demonstrated that restriction enzyme digestion is a rapid method of analyzing these samples.
FIG. 4.
Digestion of genotype 1 RT-PCR products with Alw26I. Restriction enzyme analysis of the 5′ UTR was performed for a subset of samples that were genotype 1 by LiPA but that could not be subtyped by LiPA. 5′ UTR RT-PCR products were subjected to restriction enzyme digestion with Alw26I in order to differentiate between a G residue at position −99 (subtype 1b pattern) and an A residue at this position (subtype 1a pattern). Upper panel, diagram showing the position of Alw26I restriction sites within the 340-bp 5′ UTR RT-PCR product; bottom panel, 2% ethidium bromide-stained agarose gel of restriction enzyme products. The contents of the lanes are as follows (the lane numbers represent the unnumbered lanes from left to right, respectively): lanes 1 and 12, molecular weight markers; lane 2, subtype 1b control demonstrating 209- and 91-bp bands; lane 3, subtype 1a control demonstrating a 300-bp band; lanes 4 and 8, mixtures of subtype 1a and subtype 1b control RT-PCR products at 1:1 and 1:4 ratios, respectively; lanes 5 to 7 and 9 to 11, genotype 1 samples that were unsubtypeable by LiPA and that were shown (Fig. 2) to have both an A residue and a G residue at position −99. Sample numbers at the bottom correspond to those in Fig. 2.
The patterns observed for HCV samples with both an A residue and a G residue at position −99 could potentially be explained by the presence of a mixed subtype 1a and 1b infection. Mixing of subtype 1a and subtype 1b controls at a 1:1 ratio also demonstrated a heterogeneous pattern by Alw26 I digestion, similar to the pattern for the samples that were type 1 but unsubtypeable by LiPA. However, at a subtype 1a to subtype 1b ratio of 1:4, the uncut pattern for subtype 1a was very difficult to discern (Fig. 4). We also analyzed artificially mixed subtypes by LiPA and demonstrated a mixed LiPA 1, 2, 3, 4, 5, 6 banding pattern with a 1:1 ratio of subtype 1a to subtype 1b; however, with a 1:4 ratio the pattern for subtype 1a was very difficult to discern, and with a 1:8 ratio only the pattern for subtype 1b could be detected (data not shown).
Distribution of HCV genotypes among samples analyzed by LiPA versus NS-5b sequence analysis.
A total of 247 samples recovered from 1996 to 1998 were analyzed by NS-5b sequence analysis by using the algorithm described above. A total of 1,035 samples recovered from 1998 to 2001 were analyzed by LiPA. Figure 5 shows a comparison of the distribution of genotypes obtained in our laboratory by the two methods. Samples that could not be typed by RT-PCR were eliminated from the analysis: for samples that were known to be HCV positive by either branched DNA testing or RT-PCR, there was a 2.9% RT-PCR failure rate for LiPA and a 1.4% failure rate for sequence analysis of the NS-5b region.
The total distributions of the genotypes were very consistent between the two methods. However, there was a marked difference in the distribution of subtype 1a versus subtype 1b. The distribution by NS-5b sequence analysis was 64.4% subtype 1a and 16.6% subtype 1b. In contrast, LiPA analysis resulted in an almost equal distribution of subtype 1a versus subtype 1b (35 versus 30%). Thus, LiPA identified samples as subtype 1b at a 13% greater frequency than NS-5b sequence analysis did. Of the samples that were HCV genotype 1, LiPA identified samples as subtype 1b with a 21% greater frequency. In addition, by LiPA analysis 33 samples (3.2%) were untypeable and 82 isolates (7.9%) were called type 1 but could not be further subtyped. A total of 59 samples (5.7%) were genotype 1, unsubtypeable with band pattern 1, 2, 3, 4, 5, 6 and likely had both an A residue and a G residue at position −99. By subsequent NS-5b sequence analysis, 4 of 4 of the untypeable samples and 15 of 15 of the type 1, unsubtypeable samples were shown to be HCV genotype 1a (data not shown).
DISCUSSION
It has been suggested that interrogation of the HCV 5′ UTR, although less heterogeneous than other regions of the viral genome, correlates well with and is more sensitive than interrogation of other genomic regions of HCV and is sufficient for determining the HCV genotype (10, 15, 17, 36, 37). Although direct sequencing is more time-consuming and labor-intensive than LiPA, we have not experienced the failure rates that have been reported by other investigators for sequencing of the NS-5b region (17). Analysis of the 5′ UTR by LiPA with the INNO-LiPA HCV II kit provides a fast and easy method for determination of the HCV genotype. Although the 5′ UTR is less polymorphic, the results obtained by LiPA correlate well with those obtained by sequence analysis of the NS-5b region for determination of the genotype, with 100% concordance in this study. However, several reports have indicated that analysis of the 5′ UTR cannot accurately differentiate HCV subtypes (5, 8, 17, 22, 32, 36, 37, 39, 40, 45). This is likely because of the relative lack of heterogeneity of the 5′ UTR.
Consistent with the previous reports, in a direct comparison of 63 samples in this study, LiPA misclassified ∼20% of HCV subtype 1a isolates as subtype 1b (Table 1). Thus, LiPA tends to overcall genotype 1 samples as subtype 1b. Sequencing of both the 5′ UTR and the NS-5b gene for the isolates in six of these samples with discrepant results indicated that the isolates were clearly genotype 1a by analysis of 222 bp of NS-5b gene, but because they had a G nucleotide at position −99 they were genotyped as subtype 1b by LiPA. Consistent with other reports (17, 45), we have also identified by NS-5b gene sequencing a genotype 1b isolate that had an A residue at position −99 but that was misclassified as type 1a by LiPA analysis (data not shown). These results could be explained by (i) a mixed infection of subtype 1a and 1b isolates; (ii) infidelity of the PCR introduced by Taq polymerase, although this is unlikely to occur consistently at a single nucleotide; (iii) recombination between different HCV subtypes; or (iv) an A/G polymorphism that may exist at nucleotide −99 in HCV isolates. In the last scenario, either an A residue or a G residue, or both nucleotides, could be present in subtype 1a or subtype 1b isolates. This would invalidate the use of this single nucleotide to differentiate subtype 1a and subtype 1b isolates.
In addition, 10 genotype 1 isolates that could not be subtyped by LiPA and that had the band pattern 1, 2, 3, 4, 5, 6 had both an A residue and a G residue in the nucleotide at position −99 (Fig. 2 to 4) but were clearly genotype 1a by NS-5b sequence analysis. These results could be explained by a mixed infection with subtypes 1a and 1b or by an A/G sequence polymorphism at position −99. Although we cannot rule out the possibility that these samples represent mixed infections with genotypes 1a and 1b, 5′ UTR restriction fragment length polymorphism analysis and LiPA evaluation of artificial mixtures of isolates indicate that there would have to be an approximately equal mix of types 1a and 1b to obtain the patterns observed (Fig. 4; see also Results). A sequence polymorphism is a more likely explanation, as we would have expected to identify the isolates in at least some of these samples as genotype 1b by sequencing of the NS-5b region if they were truly mixed infections with types 1a and 1b in approximately equal proportions. To our knowledge, this is the first report that HCV genotype 1a isolates from a single patient sample can have both an A residue and a G residue at position −99. The isolates in a total of 59 of 1,035 samples tested by LiPA (5.7%) gave the band pattern 1, 2, 3, 4, 5, 6 and likely have both an A residue and a G residue at this position. The data presented here are consistent with what would be expected if the A/G at nucleotide at position −99 was a sequence polymorphism rather than segregating with a particular subtype: either an A residue or a G residue can be present at nucleotide −99 in both genotype 1a and genotype 1b isolates, and in some cases, both nucleotides can be present in a single genotype as part of the group of quasispecies present in an infected patient.
The hypothesis that the A/G at position −99 is a sequence polymorphism in the HCV genome that cannot differentiate between subtypes 1a and 1b is further supported by the genotype distribution in our laboratory of the isolates in over 200 samples analyzed by NS-5b sequencing from 1996 to 1998 and over 1,000 samples analyzed by LiPA from 1998 to 2001 (Fig. 5). The distribution in our laboratory of subtype 1a versus subtype 1b isolates was 64 and 17%, respectively, by NS-5b sequence analysis. This distribution is consistent with those in reports of studies from various regions throughout the United States, which have indicated that ∼60 and ∼20% of U.S. isolates are genotypes 1a and 1b, respectively (43). In contrast, the distributions of subtype 1a versus subtype 1b isolates by LiPA analysis were 35 and 30%, respectively. Thus, there was almost an equal distribution of subtype 1a versus subtype 1b isolates by LiPA, consistent with an inability of the A/G at position −99 to distinguish genotype 1a from genotype 1b. Interestingly, these distribution data are consistent with those in reports of other studies conducted in the United States that have used the 5′ UTR for genotyping (21, 22).
Although we cannot rule out the possibility that the relative incidence of type 1b has increased dramatically at our institution in the past 3 years, this is unlikely, as the population seen over that period was very consistent. In addition, distribution data for over 1,000 samples analyzed by LiPA in our laboratory over the past 3 years indicate that ∼30% of genotype 1a isolates were likely misclassified by LiPA as either genotype 1b or type 1, unsubtypeable (Fig. 5). These data correlate well with those obtained when the results of LiPA and NS-5b sequencing were compared in parallel (Table 1). Interestingly, these data are consistent with those in the original paper by Stuyver et al. (39) describing the use of reverse hybridization in the 5′ UTR for HCV genotyping. Of 10 prototype 1a and 1b sequences used to define reverse hybridization probes for this assay, only 70% had the pattern of an A residue for type 1a and a G residue for type 1b at position −99. Of the type 1a isolates, 30% (one of three) had a G residue at this position and would have been misclassified as type 1b; similarly, of the type 1b isolates, 30% (two of seven) had an A residue at this position and would have been misclassified as type 1a. Despite mounting evidence that analysis of the 5′ UTR does not accurately differentiate HCV subtypes, the INNO-LiPA HCV kit II provides interpretation criteria for multiple subtypes of HCV, and many laboratories use 5′ UTR analysis for determination of HCV subtypes.
In conclusion, LiPA allows the easy and rapid determination of HCV genotypes but fails to determine the subtypes of isolates in many samples and misclassifies a significant percentage of type 1a isolates as type 1b. These isolates are unlikely to be clinically significant, as most studies have identified clinical differences only between those infected with type 1 isolates and those infected with non-type 1 isolates. However, these findings have implications for epidemiologic studies, as well as for studies investigating clinical differences between patients infected with HCV genotypes 1a and 1b (11). If 5′ UTR analysis overrepresents genotype 1b or cannot differentiate between genotypes 1a and 1b, any clinical difference between patients infected with genotypes 1a and 1b will be diluted in outcome studies that interrogate the 5′ UTR for genotyping. Indeed, many studies in both the United States and Europe that have reported differences in outcomes between patients infected with genotypes 1a and 1b have used the NS-5 or core regions to define the genotype (7, 25, 28, 42, 43). On the other hand, several studies that have used the 5′ UTR for genotyping have failed to find a difference in clinical outcomes between type 1a- and type 1b-infected patients (21, 22, 24). Thus, it is possible that some of the differences in studies of the outcomes of genotype 1b infections are due to the method of genotyping used. We conclude that 5′ UTR analysis (by LiPA analysis or other methods) is sufficient for determination of the HCV type and can be used for clinical purposes to differentiate type 1 from non-type 1 isolates, but determination of subtype should rely on analysis of more heterogeneous regions of the HCV genome.
Acknowledgments
We thank Tim Oury and Obaid Shakil for critical reading of the manuscript.
REFERENCES
- 1.Alberti, A., L. Chemello, F. Noventa, L. Cavalletto, and G. D. Salvo. 1997. Therapy of hepatitis C: re-treatment with alpha interferon. Hepatology 26:137S-142S. [DOI] [PubMed]
- 2.Alter, H. J., R. H. Purcell, J. W. Shih, M. Melpolder, M. Houghton, Q.-L. Choo, and G. Kuo. 1989. Detection of antibody to hepatitis C in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N. Engl. J. Med. 321:1494-1500. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26(Suppl. 1):62S-65S. [DOI] [PubMed]
- 4.Amoroso, P., M. Rapicetta, M. E. Tosti, A. Mele, E. Spada, S. Buonocore, G. Lettieri, P. Pierri, P. Shionne, A. R. Ciccaglione, and L. Sagliocca. 1998. Correlation between virus genotype and chronicity rate in acute hepatitis C. J. Hepatol. 28:939-944. [DOI] [PubMed] [Google Scholar]
- 5.Andonov, A., and R. K. Chaudhary. 1995. Subtyping of hepatitis C virus isolates by a line probe assay using hybridization. J. Clin. Microbiol. 33:254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belli, L. S., E. Silini, A. Alberti, G. Bellati, C. Vai, E. Minola, G. Rondinara, L. de Carlis, M. Asti, D. Forti, and G. Ideo. 1996. Hepatitis C virus genotypes, hepatitis, and hepatitis C virus recurrence after liver transplantation. Liver Transplant. Surg. 2:200-205. [DOI] [PubMed] [Google Scholar]
- 7.Bruno, S., E. Silini, A. Crosignani, F. Borzio, G. Leandro, F. Bono, M. Asti, S. Rossi, A. Larghi, A. Cerino, M. Podda, and M. U. Mondelli. 1997. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a prospective study. Hepatology 25:754-758. [DOI] [PubMed] [Google Scholar]
- 8.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 9.Cha, T.-A., J. Kolberg, B. Irvine, M. Stempien, E. Beall, M. Yano, Q. L. Choo, M. Houghton, G. Kuo, J. H. Han, et al. 1991. Use of a signature nucleotide sequence of hepatitis C virus for detection of viral RNA in human serum and plasma. J. Clin. Microbiol. 29:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, S.-W., F. McOmish, E. C. Holmes, B. Dow, J. F. Peutherer, E. Follett, P. L. Yap, and P. Simmonds. 1992. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J. Gen. Virol. 73:1131-1141. [DOI] [PubMed] [Google Scholar]
- 11.Charlton, M. 2000. Genotype 1b and severity of posttransplant recurrence of hepatitis C infection—unconvictable felon or wrongly accused? Liver Transplant. 6:243-249. [DOI] [PubMed] [Google Scholar]
- 12.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 13.Choo, Q.-L., K. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, G. L., R. Esteban-Mur, R. Vinod, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M.-H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 15.Dixit, V., S. Quan, P. Martin, D. Larson, M. Brezina, R. Dinello, K. Sra, J. Y. N. Lau, D. Chien, J. Kolberg, et al. 1995. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J. Clin. Microbiol. 33:2978-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto, N., A. Takada, T. Nakao, and T. Date. 1990. There are two major types of hepatitis C virus in Japan. Biochem. Biophys. Res. Commun. 170:1021-1025. [DOI] [PubMed] [Google Scholar]
- 17.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon, F. D., J. J. Poterucha, J. Germer, N. N. Zein, K. P. Batts, J. B. Gross, Jr., R. Wiesner, and D. Persing. 1997. Relationship between hepatitis C genotype and severity of recurrent hepatitis C after liver transplantation. Transplantation 63:1419-1423. [DOI] [PubMed] [Google Scholar]
- 19.Han, C. J., H. S. Lee, H. S. Kim, J. H. Choe, and C. Y. Kim. 1997. Hepatitis C virus genotypes in Korea and their relationship to clinical outcome in type C chronic liver diseases. Korean J. Intern. Med. 12:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 21.Lau, J. Y. N., G. L. Davis, L. E. Prescott, G. Maertens, K. L. Lindsay, K. Quan, M. Mizokami, and P. Simmonds. 1996. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in the United States. Ann. Intern. Med. 124:868-876. [DOI] [PubMed] [Google Scholar]
- 22.Lau, J. Y. N., M. Mizokami, J. A. Kolber, G. L. Davis, L. E. Prescott, T. Ohno, R. P. Perrillo, K. L. Lindsay, R. G. Gish, K.-P. Qian, M. Kohara, P. Simmonds, and M. S. Urdea. 1995. Application of six hepatitis C genotyping systems to sera from chronic hepatitis C patients in the United States. J. Infect. Dis. 171:281-289. [DOI] [PubMed] [Google Scholar]
- 23.McHutchinson, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M.-H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 24.Naoumov, N. V., S. Chokshi, E. Metivier, G. Maertens, P. J. Johnson, and R. Williams. 1997. Hepatitis C virus infection in the development of hepatocellular carcinoma in cirrhosis. J. Hepatol. 27:331-336. [DOI] [PubMed] [Google Scholar]
- 25.Nousbaum, J. B., S. Pol, B. Nalpas, P. Landais, P. Berthelot, C. Brechot, and the Collaborative Study Group. 1995. Hepatitis C virus type 1b (II) infection in France and Italy. Ann. Intern. Med. 122:161-168. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto, H., S. Okada, Y. Sugiyama, S. Yotsumoto, T. Tanaka, H. Yoshizawa, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1990. The 5′ terminal sequence of the hepatitis C virus genome. Jpn. J. Exp. Med. 60:167-177. [PubMed] [Google Scholar]
- 27.Pageaux, G. P., J. Ducos, A. M. Mondain, V. Costes, M. C. Picot, P. F. Perrigault, J. Domergue, D. Larrey, and H. Michel. 1997. Hepatitis C virus genotypes and quantitation of serum hepatitis C virus RNA in liver transplant recipients: relationship with severity of histological recurrence and implications in the pathogenesis of HCV infection. Liver Transplant. Surg. 3:501-505. [DOI] [PubMed] [Google Scholar]
- 28.Pistello, M., F. Maggi, L. Vatteroni, N. Cecconi, F. Panicucci, G. P. Bresci, L. Gambardella, M. Taddei, A. Bionda, M. Tuoni, and M. Bendinelli. 1994. Prevalence of hepatitis C genotypes in Italy. J. Clin. Microbiol. 32:232-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pozzato, G., M. Moretti, L. S. Croce, F. Sasso, S. Kaneko, M. Unoura, K. Kobayashi, M. Crovatto, G. Santini, and C. Tiribelli. 1995. Interferon therapy in chronic hepatitis C virus: evidence of different outcome with respect to different viral strains. J. Med. Virol. 45:445-450. [DOI] [PubMed] [Google Scholar]
- 30.Shuhart, M. C., M. P. Bronner, D. R. Gretch, L. V. Thomassen, C. F. Wartelle, H. Tateyama, S. S. Emerson, J. D. Perkins, and R. L. Carithers. 1997. Histological and clinical outcome after liver transplantation for hepatitis C. Hepatology 26:1646-1652. [DOI] [PubMed] [Google Scholar]
- 31.Silini, E., F. Bono, A. Cividini, A. Cerino, S. Bruno, S. Rossi, G. Belloni, B. Brugnetti, E. Civardi, L. Salvaneschi, and M. U. Mondelli. 1995. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology 21:285-290. [PubMed] [Google Scholar]
- 32.Simmonds, P. 1995. Variability of hepatitis C virus. Hepatology 21:570-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 34.Simmonds, P., E. C. Holmes, T.-A. Cha, S.-W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds, P., F. McOmish, P. L. Yap, S.-W. Chan, C. K. Lin, G. Dusheiko, A. A. Saeed, and E. C. Holmes. 1993. Sequence variability in the 5′ noncoding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J. Gen. Virol. 74:661-668. [DOI] [PubMed] [Google Scholar]
- 36.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap, J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053-1061. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. B., J. Mellor, L. M. Jarvis, F. Davidson, J. Kolberg, M. S. Urdea, P. L. Yap, P. Simmonds, and the International HCV Collaborative Study Group. 1995. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J. Gen. Virol. 76:1749-1761. [DOI] [PubMed] [Google Scholar]
- 38.Stuyver, L., R. Rossau, A. Wyseur, M. Duhamel, B. Vanderborght, H. Van Heuverswyn, and G. Maertens. 1993. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J. Gen. Virol. 74:1093-1102. [DOI] [PubMed] [Google Scholar]
- 39.Stuyver, L., A. Wyseur, W. Van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuyver, L., A. Wyseur, W. Van Arnhem, F. Lunel, P. Laurent-Puig, J. M. Pawlotsky, B. Kleter, L. Bassit, J. Nkengason, L. J. van Doorn, et al. 1995. Hepatitis C genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 38:137-157. [DOI] [PubMed] [Google Scholar]
- 41.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zein, N. N., J. J. Poterucha, J. B. Gross, R. H. Wiesner, T. M. Therneau, A. A. Gossard, N. K. Wendt, P. S. Mitchell, J. J. Germer, and D. H. Persing. 1996. Increased risk of hepatocellular carcinoma in patients infected with hepatitis C genotype 1b. Am. J. Gastroenterol. 91:2560-2562. [PubMed] [Google Scholar]
- 43.Zein, N. N., J. Rakela, E. L. Krawitt, K. R. Reddy, T. Tominaga, D. H. Persing, et al. 1996. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann. Intern. Med. 125:634-639. [DOI] [PubMed] [Google Scholar]
- 44.Zein, N. N., J. Rakela, J. J. Poterucha, J. L. Steers, R. H. Wiesner, and D. H. Persing. 1995. Hepatitis C genotypes in liver transplant recipients: distribution and 1-year follow-up. Liver Transplant. Surg. 1:354-357. [DOI] [PubMed] [Google Scholar]
- 45.Zeuzem, S., B. Ruster, J. H. Lee, T. Stripf, and W. K. Roth. 1995. Evaluation of a reverse hybridization assay for genotyping of hepatitis C virus. J. Hepatol. 23:654-661. [DOI] [PubMed] [Google Scholar]