Abstract
Hepatitis C virus (HCV), the causative agent of hepatitis C, frequently causes chronic infection. The mechanisms of viral persistence continue to be the object of investigation. An important aspect of HCV chronic infection is the quasispecies nature of the viral population, which has been particularly well documented in the hypervariable region 1 of the E2 glycoprotein. Recent studies show that characterization of the quasispecies diversity at the amino acid level can help to predict the outcome of HCV infection. Currently the accurate characterization of HCV quasispecies requires the cloning of PCR products, followed by the sequencing of many clones. In this study we present a new method to characterize HCV quasispecies, based on in vitro translation of the amplicons, followed by mass spectrometry analysis of the resulting peptide mix. The assay was used on reference HCV samples and on clinical samples. In principle, this method could be applied to other chronic viral infections in which quasispecies play a role.
The consequences of hepatitis C virus (HCV) infection are well known and include chronic infection in as many as 85% of cases, of which up to 24% will progress to liver cirrhosis, and development of hepatocellular carcinoma (19). Ever since the demonstration of chronic infections caused by HCV, the mechanisms of viral persistence have been the subject of investigation. A related problem is the lack of protection against reinfection following successful clearance of the primary infection, in both humans and chimpanzees, in spite of the development of both humoral and cell-mediated immunity (5, 12). The quasispecies nature of HCV in a given host, i.e., the presence of several distinct, but closely related mutants of HCV that are constantly changing due to the low fidelity of the RNA polymerase (1, 11), may contribute to HCV pathogenesis, for example, through generation of immune escape mutants. In this regard, the 31-amino-acid region at the N terminus of the E2 glycoprotein, referred to as hypervariable region 1 (HVR1), has been particularly well studied, because most mutations accumulate in this domain. Interestingly, mutations in HVR1 introduced significant changes in its predicted secondary structure (20). In addition, HVR1 has been shown to contain epitopes for neutralizing antibodies (6). A convincing demonstration of the role of quasispecies diversity of HVR1 in HCV pathogenesis was published recently by Farci and coworkers (7). In that study it was shown that the outcome of HCV infection is predicted by the change in diversity of HVR1 amino acid sequences at the time of seroconversion, an increase in HVR1 diversity being associated with chronicity. Furthermore, a larger number of nonsynonymous mutations occurring in HVR1 were documented in cases of progressive hepatitis. This is consistent with the demonstration that some mutants in HVR1 are indeed immune escape mutants (8). The greater variation in amino acid sequence of the HVR1 in the viral population and the progression of disease are thought to reflect the inability of the immune system to contain viral variation (7).
In practice, genetic changes in HCV must be demonstrated by PCR amplification of the HVR1-coding region of the HCV genome from samples obtained before and after seroconversion, followed by cloning and sequencing of at least 10 clones from which the corresponding amino acid sequences are determined (7). This procedure would entail enormous logistical problems for a clinical microbiology laboratory. In order to address this issue, we have developed a more-streamlined mutation detection method for the HVR1 region of HCV that uses matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric analysis of in vitro-synthesized peptides as previously described (10). Amplicons are generated by reverse transcription (RT)-PCR using RNA template extracted from serum samples, and these amplicons are in turn used as a template to synthesize tagged peptides in a coupled in vitro transcription-translation reaction. The tagged test peptides are then purified using the tag prior to mass determination by mass spectrometry. Variations in the nucleic acid sequence of the virus translate into mass shifts in the encoded peptides. A major advantage of this method for quasispecies analysis, in addition to ignoring silent mutations that cannot affect immunogenicity, is that MALDI-TOF mass spectrometry can detect mutant peptides present at a low frequency in a mixture, which is not possible by direct DNA sequencing of the amplicon, hence the need for the tedious process of sequencing multiple cloned PCR products
MATERIALS AND METHODS
HCV strains and plasmids.
The HCV strain H-77 (8) in the form of a serum aliquot; the HCV infectious clone pCV-H77C of strain H-77, genotype 1a (25); and the infectious clone pCV-J4L6S of strain J4, genotype 1b (26), were obtained from R. H. Purcell and Jens Bukh, Hepatitis Viruses Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Primers.
Primers were designed to bracket the segment encoding the HVR1, targeting regions conserved between the sequences of pCV-H77C and pCV-J4L6S. Primer pairs were designed for performing nested PCR. The outer pair consisted of primer HCPEP-1 (5′GGTTCTGATTGTGCTGCTACTATTTGC 3′), whose sequence is homologous to the end of the region coding for the C terminus of E1, just ahead of the E1-E2 junction, and HCPEP-2 (5′ CTATTGATGTGCCAACTGCCG 3′), whose sequence is homologous to the segment of the E2 gene just after the end of the HVR1 coding segment. The outer pair generates an amplicon of 161 bp. Several different primers were used for the second round of PCR. The sense primer HCFLAG-3 (Fig. 1) consisted of a restriction site upstream of a T7 promoter consensus sequence; filler DNA to allow for ribosomal scanning; a Kozak consensus sequence; and a start codon in frame with the segment encoding for the Flag tag (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) followed by a segment (in frame) homologous to HCV sequence, overlapping with HCPEP-1, and internal to it by 3 nucleotides. The antisense primer HCFLAG-4 contained a stretch of 15 Ts to generate a poly(A) tail during transcription, a stop codon in frame with a segment encoding the Flag tag and in frame with a segment homologous to HCV sequence, overlapping with HCPEP-2 and internal to it by 5 nucleotides (Fig. 1). HCFLAG-3 and HCFLAG-4 generate an amplicon of 257 bp. For both HCFLAG-3 and HCFLAG-4, versions of these primers without the Flag coding segment were also synthesized and used in some experiments. Finally, primer HCFLAGALA is a modification of HCFLAG-3, containing a codon for Ala between the Met codon and the Flag coding segment (Fig. 1).
FIG. 1.
Some of the primers used to generate amplicons suitable for in vitro translation. Primer HCFLAG-3 is a sense primer containing an EcoRI restriction site upstream of a T7 promoter core sequence, followed by filler DNA (including three Gs immediately after the T7 core sequence) to allow for ribosomal scanning, a Kozak consensus sequence, a Flag tag (DYKDDDDK) coding region, and a region homologous to HCV sequence, in frame with the Flag coding region. Primer HCFLAG-4 is an antisense primer containing a stretch of 15 Ts to generate a poly(A) tail at the time of transcription, a stop codon in frame with a Flag coding region, and a region homologous to HCV sequence. Primer HCFLAGALA is a modified HCFLAG-3 with an additional codon encoding for alanine.
Synthetic peptide.
The synthetic peptide MVLLLFAGVDAETHVTGGNAGRTTAGLVGLLTPGAKQNIQLINTNGSWHIDYKDDDDK was obtained from the Peptide Synthesis Facility, Advanced Protein Technology Center, Hospital for Sick Children. The sequence of the peptide corresponds to that expected from the translation of an amplicon obtained from the infectious clone pCV-H77C, with primers encoding for the Flag tag at the C terminus only.
PCR.
When using plasmid DNA as a template, PCR with the outer primer pair was performed in a total volume of 100 μl and included 50 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, 2.5 mM MgCl2, 0.5 μl of AmpliTaq Gold (Applied Biosystems Inc., Framingham, Mass.), and the buffer supplied by the manufacturer. Reactions were performed in thin-walled PCR tubes (Stratagene), overlaid with 100 μl of mineral oil, in a Robocycler-40 thermal cycler (Stratagene) with the following parameters: one cycle consisting of denaturation at 95°C for 10 min, annealing at 50°C for 1 min, and elongation at 72°C for 1 min 30 s, followed by 35 cycles with denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and elongation at 72°C for 1 min 30 s. Nested PCR with the inner pair was performed by transferring 10% of the first PCR mix into a new PCR mix prepared essentially as described above, except for the use of an inner pair of primers and adjustment of the buffer and MgCl2, taking into account the contributions from the first round. Cycling parameters were as described above, except for the annealing temperature which was changed to 67°C.
Precautions against PCR contamination.
PCR reagents were prepared before each assay in a master mix that was then aliquoted. The preparation of the master mix, the extraction of the DNA and addition of the template to the PCR, and the thermal cycling were performed in three different, well-separated rooms, each with their dedicated set of micropipettors and gowns. General precautions against contamination, including systematic use of aerosol-barrier protected pipette tips, frequent changes of gloves, and frequent decontamination of surfaces with UV light and sodium hypochlorite, were strictly adhered to.
RT-PCR.
HCV RNA was extracted from H77 serum using the Trizol reagent (Life Technologies) as described previously (21). The RNA pellet was resuspended in 10 mM dithiothreitol and 5% (vol/vol) RNasin (20 to 40 U/μl; Promega) in nuclease-free, double-distilled water (ddH2O). The RNA pellet was heated at 65°C for 2 min and put on ice; 10.5 μl of an RT mix consisting of 0.5 μl of RNasin, 2 μl of 10× PCR buffer, 2 μl of 25 mM MgCl2, 2 μl of deoxynucleoside triphosphate (10 mM each), 3 μl of 10 μM HCPEP-2 primer, and 1 μl of avian myeloblastosis virus reverse transcriptase (5 to 10 U/μl, Promega). The reaction mixture was incubated at 42°C for 1 h and then added in totality to a PCR mix consisting of 8 μl of 10× PCR buffer, 8 μl of 25 mM MgCl2, 5 μl of each primer stock solution (10 μM) from the outer pair, 0.5 μl of AmpliTaq Gold, and 53.5 μl of ddH2O. Nested PCR was then set up and performed as described above.
In vitro translation.
In vitro translation was performed in a reticulocyte lysate system by transferring 7 μl of the second-round PCR mixture to 40 μl of master mix of the TNT T7-Quick for PCR DNA (Promega), to which were added 1 μl of 1 mM methionine and 2 μl of ddH2O, and incubating the reaction at 30°C. Incubation times of 10, 30, 60, and 90 min were studied.
Magnetic bead preparation.
Biotinylated anti-Flag antibody M2 (Sigma) was bound to magnetic beads coated with streptavidin (Dynabeads M-280; Dynal), following manufacturer's recommendations, by mixing 2.4 mg of magnetic beads with 0.1 mg of antibody. The magnetic bead-antibody preparation was resuspended in 200 μl of phosphate-buffered saline with 0.1% bovine serum albumin and kept at 4°C.
Peptide purification.
Peptide purification was performed by adding 10 μl of the magnetic bead-antibody preparation to the in vitro-translation reaction and incubating 10 min at room temperature. The beads were then separated with a magnet, washed 3 times with ammonium bicarbonate 25 mM and three times with ddH2O. Finally, beads were resuspended in 20 μl of 0.1% trifluoroacetic acid to elute the bound peptides and then were removed with a magnet. The eluted peptides were recovered in a microcentrifuge tube and lyophilized in a vacuum centrifuge (Speed-Vac). The purified peptide were then resuspended in 3 μl of an α-cyano-4-hydroxycinnamic acid-saturated solution in 50% acetonitrile and 0.1% trifluoroacetic acid, prior to MALDI-TOF analysis.
MALDI-TOF mass spectrometry.
Mass spectrometry was performed at the Mass Spectrometry Laboratory, MMRC, Faculty of Medicine, University of Toronto. MALDI-TOF mass spectrometry analyses were carried out by using an Applied Biosystems Voyager-DE STR mass spectrometer (Applied Biosystems Inc.), equipped with a pulsed UV nitrogen laser (wavelength, 337 nm; 3-ns pulse) and a dual microchannel plate detector. For protein molecular weight determination, mass spectra were acquired in linear-DE mode, acceleration voltage was set to 20 kV, grid voltage was set at 94% of the acceleration voltage, guide wire voltage was set at 0.050%, delay time was set at 175 ns, and the low mass gate was set at 1,000 Da. The protonated ions of a mixture of peptides (molecular mass, 1,294.69 to 5,734.59 Da) were used for external calibration. For sample analysis, 1.5 μl of sample solution was applied on the MALDI sample plate. Mass spectra were recorded after evaporation of the solvent and processed using Data Explorer software for data collection and analysis.
Genotyping of HCV from clinical samples.
RNA was extracted from serum samples as described above. A segment of the HCV genome at the Core-E1 junction was amplified by nested RT-PCR with the primer pairs 493S_H77-978R_H77 and 502S_H77-975R_H77 as described previously (18). The amplicon was sequenced using the primer 502S_H77 as the sequencing primer. This was performed by the DNA Sequencing Facility, Center for Applied Genomics, Hospital for Sick Children. The genotype was then determined by phylogenetic analysis, comparing the sequence with that of reference strains (18). This was performed using the Clustal X for Windows (version 1.18) program (22) and the Treeview for Windows (version 1.5.2) program (14).
RESULTS
Peptide purification and MALDI-TOF analysis.
Our initial design was to have peptides synthesized with the Flag tag at the C termini, unlike the earlier work done on tagged, in vitro-synthesized peptides that used an amino-terminal tag (10). A C-terminal tag was used to avoid the possibility of purifying peptides synthesized from mRNA that is not full length. The disadvantage of this approach is that C-terminally tagged peptides cannot be used to detect mutations that result in premature truncation of the peptide. However, the HCV genome has a single large open reading frame that encodes a polyprotein, and any premature truncation would generate a replication-defective virus. The present assay is designed to detect mutations resulting in amino acid substitutions; thus, a C-terminal tag is acceptable.
To test our purification method, a synthetic peptide with a predicted mass of 6,125 Da was made whose sequence was derived from residues of the pCV-H77C infectious clone fused to a C-terminal Flag tag. After mixing either 100 or 10 pmol of the peptide in either phosphate-buffered saline or TNT-T7 Quick master mix, we could detect the peptide by mass spectrometry following magnetic bead purification (data not shown). Even after incubation for 90 min at 30°C in the TNT mix, the peptide could still be recovered; no cleavage products were observed.
Optimization of in vitro transcription-translation.
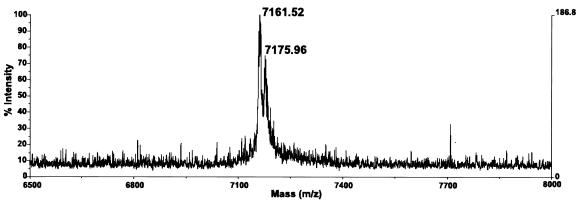
We used a PCR product obtained from the pCV-H77C infectious clone that should generate the same peptide as the synthetic peptide described above. We were unable to recover a peptide with the expected mass of 6,125 Da, although a peptide of approximately 2,394 Da was consistently recovered in these experiments that was not present in negative controls lacking template. This was interpreted to be the result of a cleavage sustained by the nascent peptide to generate the Flag tagged C-terminal fragment IQLINTNGSWHIDYKDDDDK, which has a predicted mass of 2,391 Da. Experiments with primers designed to yield a peptide with a Flag tag at the N terminus were even less successful in that we could not retrieve any peptides. However, when using the primer pair HCFLAG-3 and HCFLAG-4, designed to yield a peptide with the Flag tag at both termini, we consistently recovered a peptide of approximately 7,162 Da (Fig. 2). We found no improved yield by incubating longer than 10 min. The mass observed was approximately 42 Da more than the 7,120 Da predicted from the pCV-H77C sequence, the two Flag tags, and the amino-terminal methionine. The precision of the mass spectrometer used in these experiments was expected to be within 0.1%. This difference of 42 Da would be well accounted for by the expected N-terminal acetylation of the peptide (16, 23). Using these primers, we could also obtain a peptide of the expected mass (including acetylation) with an amplicon obtained from the pCV-J4L6S plasmid (data not shown). In several of these experiments, a second peak approximately 16 Da apart from the first peak was observed (Fig. 2), corresponding, in all likelihood, to oxidized methionine (17).
FIG. 2.
Mass spectrometry analysis of peptides obtained by translation of an amplicon obtained from clone pCV-H77C with primers HCFLAG-3 and HCFLAG-4. Based on the sequence of pCV-H77C, the predicted mass of the resulting N-terminal acetylated peptide should be 7,162 Da. In several experiments, as illustrated here, a second peak approximately 16 Da apart was observed and was interpreted as resulting from oxidation of methionine.
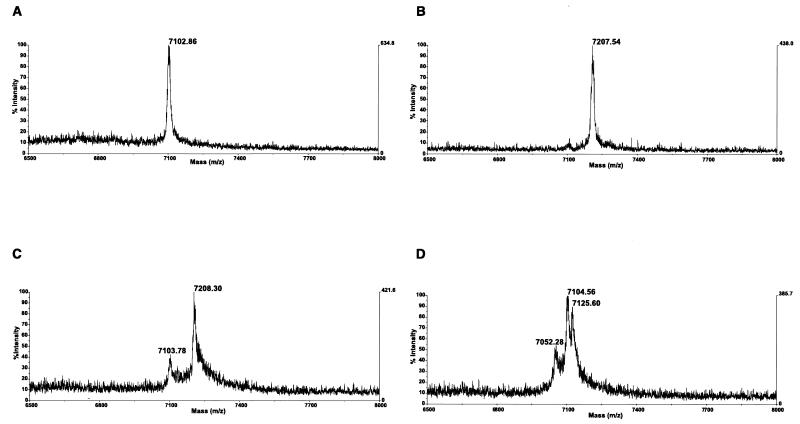
The 5′ primer HCFLAGALA was then used to replace HCFLAG-3, and was designed so that the synthesized peptide would begin by the sequence Met-Ala, ensuring the posttranslational cleavage of methionine and the acetylation of the residual peptide (16). Experiments with amplicons obtained with this primer from the pCV-H77C plasmid confirmed this hypothesis: we consistently recovered peptides with a mass of approximately 7,102 Da which is the mass predicted from the amino acid sequence encoded by the amplicons, followed by methionine cleavage and acetylation (Fig. 3A). In contrast to the experiments presented above, however, we obtained our best results after an incubation of 60 min at 30°C, suggesting that methionine cleavage, which occurs before completion of the nascent peptide (16), delayed the peptide synthesis. In agreement with our hypothesis concerning N-terminal methionine oxidation, the extra peak having an additional 16 Da was no longer observed when alanine was used to program the removal of the N-terminal methionine (Fig. 3A).
FIG. 3.
Mass spectrometry analysis of peptides translated from amplicons obtained with primers HCFLAGALA and HCFLAG-4. (A) Peptide obtained from template pCV-H77C. The predicted mass of the peptide, after methionine cleavage and N-terminal acetylation, is 7,102 Da. (B) Peptide obtained from template pCV-J4L6S. The predicted mass, after methionine cleavage and N-terminal acetylation, is 7,206 Da. (C) Peptides obtained from mixing the amplicons of pCV-H77C and pCV-J4L6S. (D) Peptides obtained from the H-77 serum aliquot.
MALDI-TOF analysis of reference HCV reagents.
In addition to the MALDI-TOF analysis of clone pCV-H77C (Fig. 3A) we also used our procedure with the clone pCV-J4L6S. We consistently obtained peptides of the predicted mass of approximately 7,206 Da (Fig. 3B). Next, we subjected to transcription-translation and mass spectrometry a mix of amplicons obtained from the two infectious clones, and we consistently obtained peptide mixtures of the two expected masses (Fig. 3C). Lastly, we performed RT-PCR on HCV RNA from the H-77 serum aliquot, whose quasispecies population has been extremely well characterized (8). As shown in Fig. 3D, we could clearly demonstrate three peptides species with observed masses of 7,052, 7,105, and 7,126 Da, respectively.
MALDI-TOF analysis of clinical samples.
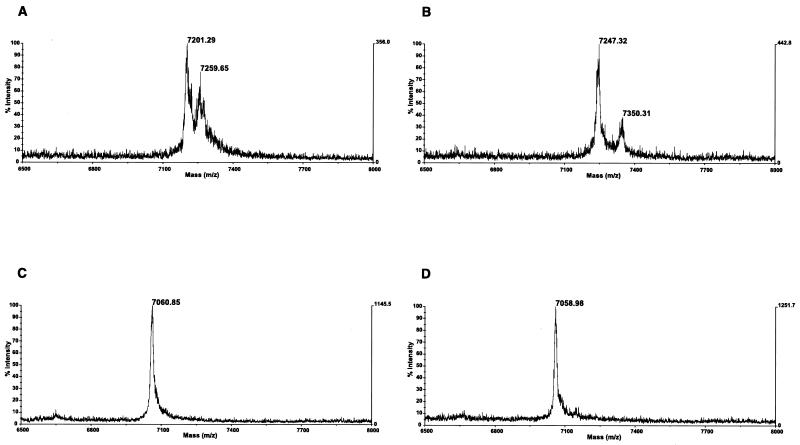
Results obtained from clinical samples are shown in Fig. 4. The first patient presented at our institution at 26 months of age; this male patient was referred for known hepatitis C infection. His mother was also suffering from chronic hepatitis C. The patient was asymptomatic, and his physical exam was unremarkable. Laboratory analysis revealed an elevated serum alanine aminotransferase (ALT) level at 111 U/liter (normal, 0 to 40 U/liter). Repeat clinical examination 6 months later revealed no changes, but the ALT level was within normal values, at 26 U/liter. This patient was infected with HCV genotype 1b. MALDI-TOF analysis on the initial serum sample demonstrated the presence of two main variants in the HCV quasispecies (Fig. 4A). Remarkably, 6 months later these had disappeared and were replaced by new variants (Fig. 4B).
FIG. 4.
Mass spectrometry analysis of clinical samples from patients with hepatitis C. (A and B) These two samples were taken 6 months apart from a patient infected with an HCV strain of genotype 1b. A complete change of the HCV population was observed during that period. (C and D) These two samples were taken 6 months apart from a patient infected with an HCV strain of genotype 1a. No changes in the HCV population could be observed.
The second patient was 21 months old at presentation in our institution. This female patient was referred for possible mother-to-infant transmission of HCV. She had been asymptomatic and her physical exam was unremarkable. However, her laboratory analysis revealed a serum ALT level of 244 U/liter. Six months later, the clinical examination remained unchanged, the ALT level remained elevated at 205 U/liter. A percutaneous liver biopsy performed at the time revealed normal liver architecture, no fibrosis, but some evidence of inflammation limited to focal collections of inflammatory cells in the sinusoids with scattered hepatocyte necrosis. This patient was infected with HCV genotype 1a. MALDI-TOF analysis on the initial sample showed a very homogeneous population, with only a single HCV variant demonstrated (Fig. 4C). Six months later, the HCV quasispecies had not changed (Fig. 4D).
DISCUSSION
In this study we have developed a procedure to evaluate the HVR1 peptide diversity in the HCV population of a patient's serum sample, without the need of cloning and sequencing of many clones. This is a conceptually straightforward application of mutation detection using MALDI-TOF (10), but several problems specific to HCV HVR1 had to be addressed, some of which were unexpected.
The power of MALDI-TOF to detect mutations relies on the changes in mass caused by amino acid substitutions. Single-amino-acid substitutions produce mass alterations ranging from 0 to 186 Da. If the peptide is short enough, most of these changes can be detected: for example in a 10-kDa peptide, all but 14 of 380 possible amino acid substitutions should be detectable with a mass accuracy of 200 ppm (10). However, the effective mass range of the MALDI instrument imposes constraints on the size of the HVR1 test sequence that is to be translated to roughly 150 bp. Unfortunately, sequence comparison between all HCV genotypes failed to reveal conserved sequence across all genotypes within such a short region around the HVR1 that could be used as primer binding sites. In this study we therefore used primers that were designed for genotype 1 and which were shown to amplify HCV strains of genotype 1a and 1b. Generalization of the method will require the design of primers specific for each genotype. These different sets of primers might conceivably be used simultaneously in a multiplex PCR to allow amplification of any HCV genotype present in the patient sample in a single reaction. Alternatively, it might be preferable to keep the reactions separated: in cases of infection by more than one genotype it would allow for monitoring the evolution of HCV quasispecies for each genotype separately. We have designed our primer pairs with the capacity to do nested PCR for increased sensitivity, but of course when working with high titer samples (or from plasmids) one round of PCR with the inner pair may suffice.
Purification of the synthesized peptides was achieved through the incorporation of the Flag tag. Since the M2 monoclonal antibody can in principle bind to the Flag sequence at either the N or C terminus, the exact location did not seem to matter and our initial design was to put the Flag at the C terminus. Experiments with a synthetic H-77 peptide confirmed that purification was achieved with our method, and no cleavage of the peptide could be observed after prolonged incubation in the reticulocyte lysate. However, with translation from an amplicon we did consistently observe a cleavage, suggesting that the nascent peptide, as it was being synthesized, was exposed to proteases. In hindsight this is not so surprising, since in order to bracket the HVR1 the amplicon must contain the segment coding for the C terminus of the E1 glycoprotein, which is highly hydrophobic and provides a transmembrane anchor to the protein, causing retention of E1 in the endoplasmic reticulum rather than channeling to the Golgi (3). Presumably, the highly hydrophobic character of the amino terminus of this peptide results in its targeting by the proteasome, which preferentially recognizes exposed hydrophobic amino acid R groups (13).
With the hydrophilic Flag tag on both ends, the peptide is apparently not exposed to protease activity, perhaps because the hydrophobic character of the amino terminus is altered by the highly charged residues of the Flag tag. We have, however, no good explanation for the poor results obtained when the Flag tag appeared only at the N terminus.
Using the HCFLAG-3 and HCFLAG-4 primers, which direct the synthesis of the Flag tag on both ends, we consistently obtained good results and obtained peptides whose masses were essentially those predicted for clone pCV-H77C (Fig. 2) and for clone pCV-J4L6S (not shown). In addition, correct masses were observed for a mix of amplicons obtained from these two clones (not shown) and three main peptide species after translation of the amplicon from the H-77 serum (not shown). In these experiments we did observe a consistent shift from the predicted mass which coincided with the predicted increase of 42 Da caused by N-terminal acetylation (16, 23). This is not a problem, since in all our experiment this enzyme mediated posttranslational modification was consistent and complete. More problematic for our purpose was the frequently observed second peak shifted by approximately 16 Da, which we attributed to incomplete and inconsistent methionine oxidation (17), a common chemical modification. To overcome this difficulty we redesigned the sense primer to direct the incorporation of an alanine between the methionine and the Flag tag. As predicted previously (16), this resulted in a posttranslational cleavage of the methionine followed by N terminal acetylation. We did not observe the 16 Da shifted second peaks when using this primer. Experiments with clones pCV-H77C and pCV-J4L6S yielded peptides of expected masses, alone or in combination (Fig. 3A to C). Finally, mass spectrometry measurements from the translated amplicon obtained from the serum aliquot of H-77 yielded at least three distinct species whose masses correspond favorably to those predicted from the sequence of the most frequent clones in the H-77 quasispecies (8): 7,102 Da for the master clone (present at a frequency of 70 per 104 clones), 7,122 for the second most common clone (with a frequency of 6 per 104 clones), and 7,056 for the third most common clone (with a frequency of 5 per 104 clones), the other 16 clones identified being present at frequencies of 4 per 104 clones or less (8). Thus, our procedure appears to give as much information as sequencing approximately 20 clones, since it detected the subspecies present at roughly 1 in 20 (mass, 7,056 Da).
The MALDI-TOF-based assay was applied to serial samples from two patients monitored for chronic hepatitis C at our institution. The first patient presented initially with elevated ALT, a marker of liver inflammation that is a correlate of immune response against HCV (2). In a follow-up sample obtained 6 months later, the quasispecies population had completely changed, in all likelihood because of effective immune selection against the initial HCV variants. Numerous studies have documented the role of immune selection in the evolution of HCV quasispecies in HVR1, including the lack of evolution in patients with agammaglobulinemia (reviewed in reference 8). It is interesting that at the time when the second sample was taken, the ALT level was normal. One may speculate that an immune response against the new variants had not yet been mounted. The second patient displayed a very homogeneous population, since only one HCV variant could be demonstrated (Fig. 4C). It is possible that this homogeneity is a consequence of the infection being acquired vertically, since in these cases the HCV population in the infant is usually very homogeneous (8). This patient also presented with elevated ALT. However, in this patient the immune response failed to eradicate the HCV variant, since 6 months later the quasispecies had exactly the same profile; at that time, the ALT level remained elevated.
Currently, the significance of decreased quasispecies diversity at the amino acid level at the time of seroconversion has been demonstrated (7). One would have expected also that since part of the anti-HCV activity of alpha interferon is immune mediated (15), diversity decreases should be predictive of treatment outcomes. This has in fact been recently demonstrated (9). At this point in time it remains unproven whether quasispecies diversity monitoring will also be of benefit in longitudinal follow-up of patients. However, the two examples presented in this study appear similar to a recent report by Curran and coworkers (4), in which patients with mild disease had an HCV population whose amino acid sequences in HVR1 changed over time, whereas patients with more severe illness had static HCV populations. Although further work will be required on this point, it is clear that the MALDI-TOF assay allows for the monitoring of HCV quasispecies in the clinical setting in a meaningful way, and is expected to contribute to a better understanding of the natural history of the disease.
In summary we have developed and streamlined a procedure based on MALDI-TOF mass spectrometry for the characterization of HCV quasispecies in HVR1. This assay is technically less demanding to perform than cloning and sequencing many clones, since in its current state it involves the translation of the (unpurified) amplicon using a commercially available kit, the purification of the peptide mix using a simple and rapid method, and the submission to MALDI-TOF analysis. The procedure described here could also be easily adapted to other chronic viral infections in which quasispecies play a role, for example the quasispecies observed in the V3 loop of the gp120 surface glycoprotein of human immunodeficiency virus (24).
Acknowledgments
We thank Y. Yang for help with the MALDI-TOF mass spectrometry and D. Mahuran for helpful discussions.
This work was supported by a grant from the Medical Research Council of Canada.
REFERENCES
- 1.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 2.Cerny, A., and F. V. Chisari. 1999. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 30:595-601. [DOI] [PubMed] [Google Scholar]
- 3.Cocquerel, L., S. Duvet, J.-C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran, R., C. L. Jameson, J. K. Craggs, A. M. Grabowska, B. J. Thomson, A. Robins, W. L. Irving, and J. K. Ball. 2002. Evolutionary trends of the first hypervariable region of the hepatitis C virus E2 protein in individuals with differing liver disease severity. J. Gen. Virol. 83:11-23. [DOI] [PubMed] [Google Scholar]
- 5.Farci, P., H. J. Alter, S. Govindarajan, D. C. Wong, R. Engle, R. R. Lesniewski, I. K. Mushawar, S. M. Desai, R. H. Miller, N. Ogata, and R. H. Purcell. 1992. Lack of protective immunity against reinfection with hepatitis C virus. Science 258:135-140. [DOI] [PubMed] [Google Scholar]
- 6.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., and R. H. Purcell. 2000. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 20:103-126. [PubMed] [Google Scholar]
- 9.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. USA 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garvin, A. M., K. C. Parker, and L. Haff. 2000. MALDI-TOF based mutation detection using tagged in vitro synthesized peptides. Nat. Bio/Technology 18:95-97. [DOI] [PubMed] [Google Scholar]
- 11.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 12.Lai, M. E., A. P. Mazzolleni, F. Argiolu, S. De Virgilis, A. Balestrieri, R. H. Purcell, A. Cao, and P. Farci. 1994. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet 343:388-390. [DOI] [PubMed] [Google Scholar]
- 13.Pacifici, R. E., Y. Kono, and K. J. A. Davies. 1993. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 268:15405-15411. [PubMed] [Google Scholar]
- 14.Page, R. D. M. 1996. Treeview: an application to display phylogenetic trees on personal computers. Computer Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 15.Pawlotsky, J.-M., G. Germanidis, P.-O Franais, M. Bouvier, A. Soulier, M. Pellerin, and D. Dhumeaux. 1999. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J. Virol. 73:6490-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polevoda, B., and F. Sherman. 2000. Nα-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 275:36479-36482. [DOI] [PubMed] [Google Scholar]
- 17.Qin, J., and B. T. Chait. 1997. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal. Chem. 69:4002-4009. [DOI] [PubMed] [Google Scholar]
- 18.Ray, S. C., R. R. Arthur, A. Carella, J. Bukh, and D. L. Thomas. 2000. Genetic epidemiology of hepatitis C virus throughout Egypt. J. Infect. Dis. 182:698-707. [DOI] [PubMed] [Google Scholar]
- 19.Seeff, L. B. 1997. Natural history of hepatitis C. Hepatology 26(Suppl. 1):21S-28S. [DOI] [PubMed]
- 20.Taniguchi, S., H. Okamoto, M. Sakamoto, M. Kojima, F. Tsuda, T. Tanaka, E. Munekata, E. E. Muchmore, D. A. Peterson, and S. Mishiro. 1993. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology 195:297-301. [DOI] [PubMed] [Google Scholar]
- 21.Tellier, R., J. Bukh, S. U. Emerson, R. H. Miller, and R. H. Purcell. 1996. Long PCR and its application to hepatitis viruses: amplification of hepatitis A, hepatitis B and hepatitis C virus genomes. J. Clin. Microbiol. 34:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins, M. R., E. Gasteiger, A. A. Gooley, B. R. Herbert, M. P. Molloy, P.-A Binz, K. Ou, J.-C. Sanchez, A. Bairoch, K. L. Williams, and D. F. Hochstrasser. 1999. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 289:645-657. [DOI] [PubMed] [Google Scholar]
- 24.Wolfs, T. F. W., J.-J. De Jong, H. Van Den Berg, J. M. G. H. Tijnagel, W. J. A. Krone, and J. Goudsmit. 1990. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependant, rapid and continuous. Proc. Natl. Acad. Sci. USA 87:9938-9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]