Abstract
A multidrug-resistant fljB-lacking Salmonella enterica serovar [4,5,12:i:−] emerged in Spain in 1997. We analyzed the genome from four strains of this serovar using a microarray containing almost all the predicted protein coding regions of serovar Typhimurium strain LT2, including the pSLT plasmid. Only a few differences from serovar Typhimurium LT2 were observed, suggesting the serovar to be Typhimurium as well. Six regions of interest were identified from the microarray data. Cluster I was a deletion of 13 genes, corresponding to part of the regulon responsible for the anaerobic assimilation of allantoin. Clusters II and IV were associated with the absence of the Fels-1 and Fels-2 prophage. Cluster III was a small group of Gifsy-1 prophage-related genes that appeared to be deleted or replaced. Cluster V was a deletion of 16 genes, including iroB and the operon fljAB, which is reflected in the serovar designation. Region VI was the gene STM2240, which appears to have an additional homologue in these strains. The regions spanning the deletions involving the allantoin operon and the fljAB operon were PCR amplified and sequenced. PCR across these regions may be an effective marker for this particular emergent serovar. While the microarray data for all isolates of the new serovar were essentially identical for all LT2 chromosomal genes, the isolates differed in their similarity to pSLT, consistent with the heterogeneity in plasmid content among isolates of the new serovar. Recent isolates have acquired a more-complete subset of homologues to this virulence plasmid. In general, microarrays can provide useful complementary data to other typing methods.
Salmonella enterica is a leading cause of food-borne gastroenteritis in humans, as well as disease in domestic and wild animals, worldwide (20). Serotyping is widely used as an epidemiological typing method for subdivision into serovars on the basis of antigenic variability at lipopolysaccharide moieties (O antigen), flagellar proteins (H antigen), and capsular polysaccharides (Vi antigen). Epidemiological surveillance allows the detection of new serovars and monitoring of the frequency of previously known serovars and the geographical and temporal distributions of serovars.
An fljB-lacking, multidrug-resistant S. enterica serovar [4,5,12:i:−] strain was detected in Spain in 1997. The selective advantage of multidrug resistance is probably one of the factors that had influenced the increased prevalence of this strain in Spain, where it became the fourth most common serovar among all Salmonella isolates from humans during the period from 1998 to 2000 (10).
There are a number of reasons to believe this new serovar to be a monophasic variant of serovar Typhimurium. The strains were lysed by the Salmonella serotype Typhimurium phage 10 (phage type DTU302). A sequence specific for Salmonella serovar Typhimurium phage types DT104 and DTU302 was present in this atypical Salmonella strain, and a serovar Typhimurium-specific IS200 fragment was present as well, suggesting that it is a monophasic Salmonella serovar Typhimurium variant (4, 11). Strains belonging to this atypical serovar were further typed using selected genetic procedures, such as arbitrarily primed PCR and pulsed-field gel electrophoresis macrorestriction analysis. These methods indicated very little genetic heterogeneity among the organisms, leading to the conclusion that they fall into a single genetic lineage or clone, which despite the lack of the fljB gene in these strains (14) seemed to be closely related with some contemporary serovar Typhimurium lineages that cause human salmonellosis.
Microarrays have been shown to be helpful for quick detection of antibiotic resistance (24), determination of virulence and pathogenicity (5, 16, 17), species determination (6), genome comparison (1, 2, 8, 18), and molecular epidemiological typing of strains (3, 9). In addition, the microarray procedure was used successfully to analyze the deletions and amplification mutations commonly found in Salmonella mutagenicity assay strains (Ames test) (21). Here, we analyze the genome of four strains which belong to the atypical fljB-lacking emergent Salmonella serovar [4,5,12:i:−] using DNA-DNA hybridization to a microarray consisting of most of the 4,596 predicted protein-coding regions (CDSs) in the genome of S. enterica serovar Typhimurium strain LT2 (18, 21).
We have characterized the gene content of the strains shared with Typhimurium LT2 and determined which genes are not present in the atypical strains, and which genes are amplified. PCR and DNA sequencing further characterized the deletions revealed by the microarray data. The results illustrate the power of genomic analysis through the use of DNA microarrays, indicate the low genetic heterogeneity between strains of the atypical Salmonella serovar [4,5,12:i:−], and support the hypothesis of a close relationship of this clone with the serovar Typhimurium.
MATERIALS AND METHODS
Bacterial strains.
The four strains investigated were all Salmonella enterica serovar [4,5,12:i:−] strains isolated at the Spanish National Salmonella Reference Laboratory, Centro Nacional de Microbiología (CNM), Madrid. They were assigned to a serovar by the slide agglutination method with commercial antiserum (Sanofi Diagnostic Pasteur, Paris, France) and antiserum prepared in-house. Strain CNM9IC was isolated in 1997 and was the first clinical isolate identified in Spain as belonging to this atypical serovar. Strains CNM3IC and CNM4IC were isolated in 1998 from pork meat and sausages and were the first food isolates associated with the outbreak. Strain CNM1404 was isolated in 2001 from feces of a patient with gastroenteritis in Spain. Wild-type S. enterica serovar Typhimurium LT2 is available from the American Type Culture Collection (ATCC 700720).
DNA extraction and probe labeling.
Genomic DNA from the bacterial strains was prepared from 5 ml of fresh overnight culture grown in Luria-Bertani broth at 37°C using the DNEasy Tissue Kit (QIAGEN, Valencia, Calif.), according to the manufacturer's instructions. Aliquots of 1.5 μg were labeled according to P. Brown's protocol (http://cmgm.stanford.edu/pbrown/protocols/4_genomic.html) with 12 μg of random hexamers (Sigma Genosys, The Woodlands, Tex.), 10 U of Klenow enzyme (New England Biolabs, Beverly, Mass.), and 2 nmol of Cy3-dCTP or Cy5-dCTP (Amersham, Piscataway, N.J.) for 16 h at 37°C in the dark. Probes were purified using the QIAquick PCR purification kit (QIAGEN) following the manufacturer's recommendations, dried in vacuum, and solubilized in 10 μl of sterile distilled water. Immediately before use the probes were resuspended in hybridization buffer containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% sodium dodecyl sulfate, and sonicated salmon sperm DNA (200 μg/ml; Stratagene, La Jolla, Calif.) and were denatured by boiling for 2 min.
Microarray construction, hybridization, image acquisition, and data analysis.
S. enterica serovar Typhimurium open reading frame (ORF) microarray construction was carried out as previously described (18, 21). PCR amplified ORFs were spotted on glass slides following the standard protocol developed by P. Brown, Stanford, Calif. (http://cmgm.stanford.edu/pbrown/protocols/). Gamma amino propyl silane-coated glass slides (Corning Incorporated, Corning, N.Y.) or SuperAmine-coated slides (TeleChem International, Sunnyvale, Calif.) were used following the manufacturer's instructions. The spotter and software (Omnigrid and Gridder 2.0) were from GeneMachines, San Carlos, Calif. The genomic DNA of each strain of Salmonella serotype [4,5,12:i:−] was labeled with Cy3 or Cy5 fluorophores, mixed with an equal amount of alternately labeled genomic DNA of Salmonella serovar Typhimurium LT2, and applied to the array. For each assay two slides (each containing triplicate arrays) were hybridized to reciprocal mixtures of Cy3- and Cy5-labeled genomic DNA to generate six data points per gene. Genomic probes were hybridized to the microarray overnight at 55°C for Corning slides or 62°C for TeleChem slides, using a hybridization chamber (TeleChem International) submerged in water and protected from light. Before scanning the slides were washed following the manufacturer's recommendations and dried by spinning at 500 rpm for 5 min at room temperature. Scans were performed on a ScanArray 5000 Laser scanner (Packard BioChip Technologies, Billerica, Mass.) using ScanArray 3.1. software. Signal intensities were quantified using QuantArray 3.0 software (Packard BioChip Technologies). Spots were analyzed by the adaptive quantification method with local background subtractions, the percentage of contribution of each spot to total signal in each channel was calculated, ratios of these percentages were identified, and the median of the six ratios per gene was recorded. Scattered data graphs with the log10 of median ratio values of CNM strains against LT2 strain were produced. Genes displaying low hybridization (lowest, 5%) signals with LT2 strain probes were excluded before graphical representation. Log values lower than −0.4 were taken as defining the absence of a gene in CNM strains. This number is more than 3 standard deviations from the mean of the ratios for a set of genes known to be shared by salmonella and three other enterobacteria (18). A log value of +0.4 was chosen to identify a gene amplification seen in all the CNM strains.
Validation of microarray data by PCR and sequencing.
Primers were designed that were complementary to the genes with positive hybridization signals on the microarray that flanked the putative gene deletions related to the allantoin-glyoxylate pathway (gene STM0516: 5′-AGCAGGCGCGTGAACAGGGC-3′; gene STM0530: 5′-GTGCGGACGTGCGCGACGAATTT-3′). The oligonucleotides were synthesized by Genset (La Jolla, Calif.). The PCR was performed in 50-μl reaction volumes using 10 ng of genomic DNA, 50 μM deoxynucleoside triphosphate, 5 μM concentrations of primers, and 0.5 U of ExTaq (TaKaRa Biomedicals, Shiga, Japan). The cycling temperatures were as follows: incubation at 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 66°C for 30 s, 72°C for 3 min, and a final incubation at 72°C for 10 min. PCR products were verified by gel electrophoresis in 1% (wt/vol) agarose (Fisher Scientific, Fair Lawn, N.J.) in 1× Tris-borate-EDTA buffer for 60 min at 100 V. Gels were stained in ethidium bromide and recorded by photography under UV light. 1 KB Plus marker (Promega, Madison, Wis.) was used as a molecular weight standard. PCR products were purified using the QIAquick PCR purification kit (QIAGEN) and cloned using the TA Cloning Kit and vector pCR 2.1. (Invitrogen, Carlsbad, Calif.). The recombinant plasmid was purified using the QIAprep Miniprep Kit (QIAGEN) and sequenced by the dideoxy chain termination procedure.
Primers complementary to the genes with a positive hybridization on the microarray that flanked the putative gene deletions of the fljAB operon (gene STM2757: 5′-GTAAGCCGGTTCATTACGCAGCC-3′; gene STM2774: 5′-CGAACTATCCAGGCACGAGCG-3′) were designed following the same procedure as described above. The oligonucleotides were synthesized by Amersham Pharmacia Biotech Inc. (Barcelona, Spain). PCR was carried out using Ready to Go pellets (Amersham Pharmacia Biotech Inc.). The PCR was performed in 25-μl reaction volumes. The cycling temperatures were as follows: incubation at 94°C for 4 min followed by 25 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 2 min, and a final incubation at 72°C for 10 min. The PCR product for the atypical strains was purified using the QIAquick PCR purification kit (QIAGEN) and sequenced. Sequencing was carried out using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems/Perkin-Elmer, Madrid, Spain).
RESULTS
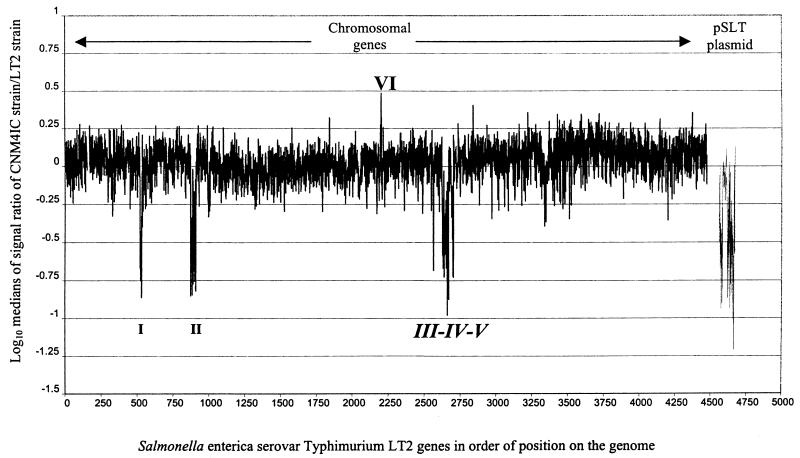
Data were collected for each of four strains of the new emergent serovar. The data for each strain were essentially identical with regard to the presence and absence of chromosomally located genes. Six genomic areas of interest were defined from the microarray data, including five clusters of genes not present on the CNM strains. One gene amplification in these strains was also observed when compared to the LT2 strain (Fig. 1). As shown in Table 1, cluster I was characterized by an absence or deletion of 13 genes (STM0517 to STM0529), corresponding to part of the regulon responsible for the anaerobic assimilation of allantoin as a nitrogen source, which is linked to the glyoxylate metabolic pathway. Cluster II was characterized by an absence of the complete Fels-1 prophage and the two adjacent genes downstream, annotated STM0928 (nanH) and STM0929, which code for a sialidase-neuraminidase and for a hypothetical protein, respectively. Cluster III was defined as a small group of Gifsy-1 prophage-related genes (STM2616 and STM2617) that were not present in the CNM strains. Cluster IV was a complete deletion of the Fels-2 prophage (STM2694 to STM2740). Downstream from this deletion, the presence of a group of 18 genes was detected in all CNM strains. At the end of this region a deletion of 16 genes was observed (cluster V), which included the operon fljAB (fljA, fljB, and hin). The fljB gene encodes the phase-2 flagellin protein, and lack of this flagellin is one of the major characteristics of this group of atypical monophasic Salmonella strains. The gene annotated STM2773, located next to the phase-2 flagellar operon and coding for a glycosyl transferase related to UDP-glucuronosyltransferase (iroB), was also deleted. Finally, an amplification or presence of additional close homologues of the gene STM2240 was observed (region VI). STM2240 codes for a putative cytoplasmic protein and is located adjacent to the sspH2 gene, the product of which is exported by the type III secretion system (TTSS) encoded by the pathogenicity island SPI-2 (19).
FIG. 1.
Quantitative data obtained from hybridization of genomic DNA of Salmonella serovar [4,5,12:i:−] CNM4IC strain versus the genome of Salmonella serovar Typhimurium LT2 and plasmid pSLT. The five gene deletions and one gene amplification are marked in roman numerals.
TABLE 1.
Deleted or amplified genomic regions in atypical monophasic Salmonella serovar [4,5,12:i:−] relative to serovar Typhimurium strain LT2
| Region | Gene no.a | No. of implicated genes | Genetic event |
|---|---|---|---|
| I | STM0517 to STM0529 | 13 | Partial deletion: allantoin-glyoxylate pathway |
| II | STM0893 to STM0929 | 37 | Deletion: complete Fels-1 prophage and two adjacent genes |
| III | STM2616 to STM2617 | 2 | Partial deletion: Gifsy-1 prophage |
| IV | STM2694 to STM2740 | 47 | Deletion: Fels-2 prophage |
| V | STM2758 to STM2773 | 16 | Deletion: fljAB operon and flanking genes |
| VI | STM2440 | 1 | Amplification: hypothetical cytoplasmic protein gene |
STM numbers correspond to the numbers in reference 18.
The DNA microarray contains 108 protein-coding regions annotated in the serovar-characteristic virulence plasmid pSLT genome. In CNM9IC, the oldest isolate, most of the plasmid genes (90%) were absent, including the spv virulence-related genes. In the more-recent isolates 40 to 60% of the pSLT genes had homologues. These results are consistent with the known heterogeneity in plasmid content among strains (14, 15).
The data include the status of all the chromosomal virulence genes previously detected by other authors (14) using PCR methods; invE/A, which is associated with invasion; stn, which is associated with the production of enterotoxin; slyA, which is associated with cytolysin; and pho, which is associated with resistance to macrophages. These genes were present in all CNM strains.
The deletions of the genes related to the anaerobic assimilation of nitrogen from allantoin and the glyoxylate pathway (cluster I) and the operon fljAB (Cluster V) were further investigated by PCR and DNA sequencing. Primers were chosen in genes adjacent to the apparent deletions. PCR across the regions generated the same size of product in all four strains and no product in LT2, as expected because the primers were too far apart in the genome unless it had the deletion (data not shown). The region spanning each deletion was sequenced in order to establish structure and features. The deletion that spans 13 of the genes related to the allantoin and glyoxylate pathway (cluster I) affected two-thirds of the gcl gene and the downstream intergenic region between the fdrA and ylbE genes. At the site of deletion there were two extra bases. Furthermore, a replacement of approximately 150 bp by a sequence not found in the GenBank database had occurred in the truncated gcl gene. The deletion affecting the fljAB operon spans 16 genes. It starts at nucleotide 228 of STM2757 with the insertion of an IS26, which is followed by the tnpA transposase gene, a repeated sequence of 14 nucleotides, and then the STM2773 gene (iroB), starting at nucleotide 564. The PCR products had sequences flanking the deletions that were identical to the sequence of serovar Typhimurium.
DISCUSSION
Most strains of Salmonella express two serologically distinct flagellar antigens. Alternating expression of two different flagellar genes, fliC and fljB, is mediated at the molecular level by an intricate mechanism which is unique to Salmonella. The two antigens were historically termed phases, and strains expressing both flagellar types are therefore called biphasic. Monophasic Salmonella could theoretically originate in two different ways. It could represent ancestral forms which did not acquire a second flagellar antigen or the necessary switching mechanism during evolution. Alternatively, it could originate as mutants of biphasic Salmonella, which have lost either the switching mechanism or the ability to express the second flagellar antigen (4).
An atypical fljB-lacking and multidrug-resistant S. enterica subsp. enterica serovar [4,5,12:i:−] emerged and spread in Spain in 1997. It was unclear whether this serovar was a new variant of the rare serovar Lagos that has an antigenic formula [4,5,12:i:1,5] similar to that of the atypical strains, but which has never been isolated in Spain, or a new variant of the very common serovar Typhimurium [4,5,12:i:1,2]. The presence of a unique sequence specific for Salmonella serovar Typhimurium phage types DT104 and DTU302, in addition to the Typhimurium serovar-specific location of copies of IS200, strongly suggested that the atypical strains are monophasic variants of serovar Typhimurium (11).
Microarray analysis of the first isolate of the atypical Salmonella serovar [4,5,12:i:−] emergent in Spain and of more-recent isolates was used to determine the genetic similarity with the serovar Typhimurium genome and also the possible evolution of this atypical serovar genome over time. All the isolates were identical in the features that distinguished them from the chromosome of LT2 on the microarray, although wide variations in the presence of homologues of plasmid-related genes were observed.
The most important deletion in the emergent serovar is the deletion of the fljAB region, which is responsible for the new serovar status. Sequencing of the deleted region indicated it to be a simple deletion with an IS26 element at the site of deletion. Transposable elements are associated with chromosomal rearrangements such as deletions, duplications, inversions, and other genetic events (13, 23). IS26 almost certainly had a role in the deletional event that generated the monophasic phenotype in these strains.
The genes required for assimilation of allantoin nitrogen and glyoxylate metabolism by Escherichia coli include a cluster of 12 ORFs (7). The atypical CNM strains showed a deletion of most of the regulon, while all other Salmonella serovar Typhimurium strains analyzed in our laboratory harbor it (data not shown). The borders of this deletion do not contain any obvious evidence of why the deletion occurred; however, it is not a simple deletion, as it includes the replacement of 150 bases nearby that have no known homologue in the GenBank database. The metabolic implications of such a deletion in the atypical Salmonella strains will require further studies. In any event, PCR across this deleted region could be a convenient marker of this emergent serovar.
Salmonella serovar Typhimurium LT2 contains at least four functional prophages, Fels-1 and -2 and Gifsy-1 and -2. Both Gifsy-1 and -2 have been implicated in the infection process (12, 22). One or more of these phages are missing from many strains of serovar Typhimurium (data not shown). The atypical CNM strains are missing both the Fels-1 and -2 prophages. More importantly, all of Gifsy-2 and almost all of the Gifsy-1 genes have homologues in the atypical strains. The occurrence of these phages in serovars other than Typhimurium has not been observed to date (18).
There are two important limitations of the method we have used in this study. First, subtle changes that involve only a few bases or replacement of genes with close homologues would not be observed by the method we have used, which requires dramatic changes in a gene, such as deletion. Second, genes not found in LT2 may be present in the CNM strains. These genes would not be observed by the method we used, as they are not located on the microarray. Guerra et al. (15) established that the atypical Salmonella serovar [4,5,12:i:−] isolated in Spain carried a class 1 integron harboring dfrA12 and aaA2 gene cassettes, and blaTEM-1, aac(3)-IV, cmlA1, and tetA genes located in large plasmids of about 140 kb (carrying spv) or 120 kb (lacking spv). The large plasmids found in these strains carry genes different from those found in the pentadrug-resistant serovar Typhimurium DT104 clone. Our results confirm that spv genes are absent in CNM9IC but present in CNM3IC, CNM4IC, and CNM1404 strains. Between 40 and 90% of the pSLT plasmid genes do not have close homologues in the CNM strains. The different isolates of the new serovar contain plasmids of different sizes; the sizes also differ from that of pSLT (14, 15). Thus, it is perhaps not surprising that they also differ in the number of homologues they share with pSLT.
Strains isolated from the emergence of the atypical serovar in Spain in 1997 and more recently were all very similar, with the exception of the differences in hybridization detected among genes of the pSLT plasmid. These data confirm the low genetic heterogeneity found by other authors using molecular epidemiological techniques when analyzing the atypical strains (14). Our data indicate that the atypical monophasic Salmonella strains are following an epidemic clonal model in terms of microbial population structure (23).
In summary, microarray technology has allowed us to determine that the genome of the atypical serovar [4,5,12:i:−] emergent in Spain contains almost all the genes present in serovar Typhimurium strain LT2, including two of four known phages. Phage is a volatile component of the genome, so the presence of phage indicates close similarity to serovar Typhimurium. Furthermore, only two additional non-phage gene clusters are absent in the atypical serovar compared to serovar Typhimurium LT2. In contrast, the other serovars within subspecies I that have been tested so far all differ from serovar Typhimurium LT2 by at least 10 clusters of genes (reference 18 and unpublished results). These observations lend further credence to the hypothesis that this atypical serovar is a variant of serovar Typhimurium. Typing of strains on arrays may contribute to understanding the epidemiology and pathology of bacterial strains because it allows genomes to be classified independent of typing of a small set of surface antigens or phage. Gene content information is more easily adapted to measuring population and evolutionary distances than other typing methods.
Acknowledgments
Javier Garaizar was the recipient of a travel grant given by the Spanish Society for Chemotherapy.
This work was supported by NIH grants AI34829 and AI43283 and the Basque Government, Spain, grant PI 1998/52.
REFERENCES
- 1.Akman, L., R. V. M. Rio, C. B. Beard, and S. Aksoy. 2001. Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of Tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J. Bacteriol. 183:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Björkholm, B., A. Lundin, A. Sillén, K. Guillemin, N. Salama, C. Rubio, J. I. Gordon, P. Falk, and L. Engstrand. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 69:7823-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnens, A., J. Stanley, I. Sechter, and J. Nicolet. 1996. Evolutionary origin of a monophasic Salmonella serovar, 9,12:l,v:−, revealed by IS200 profiles and restriction fragment polymorphisms of the fljB gene. J. Clin. Microbiol. 34:1641-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, J.-A., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusa, E., N. Obradors, L. Baldomà, J. Badía, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181:7479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, Y., J. D. Glasner, F. R. Blattner, and E. W. Triplett. 2001. Genomic interspecies microarray hybridization: rapid discovery of three thousand genes in the maize endophyte, Klebsiella pneumoniae 342, by microarray hybridization with Escherichia coli K-12 open reading frames. Appl. Environ. Microbiol. 67:1911-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrel, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echeita, M. A., A. Aladueña, S. Cruchaga, and M. A. Usera. 1999. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:− strain in Spain. J. Clin. Microbiol. 37:3425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeita, M. A., S. Herrera, and M. A. Usera. 2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:− appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 13.Gray, Y. H. M. 2000. It takes two transposons to tango. Trends Genet. 16:461-468. [DOI] [PubMed] [Google Scholar]
- 14.Guerra, B., I. Laconcha, S. M. Soto, M. A. González-Hevia, and M. C. Mendoza. 2000. Molecular characterisation of emergent multiresistant Salmonella enterica [4,5,12:i:-] organisms causing human salmonellosis. FEMS Microbiol. Lett. 190:341-347. [DOI] [PubMed] [Google Scholar]
- 15.Guerra, B., S. M. Soto, J. M. Argüelles, and M. C. Mendoza. 2001. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:−]. Antimicrob. Agents Chemother. 45:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlyshev, A. V., P. C. F. Oyston, K. Williams, G. C. Clark, R. W. Titball, E. A. Winzeler, and B. W. Wren. 2001. Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associate genes. Infect. Immun. 69:7810-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato-Maeda, M., Q. Gao, and P. M. Small. 2001. Microarray analysis of pathogens and their interaction with hosts. Cell. Microbiol. 3:713-719. [DOI] [PubMed] [Google Scholar]
- 18.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterson, and R. K. Wilson. 2001. The complete genome sequence of Salmonella enterica serovar Typhimurium LT2: features revealed by comparison to related genomes. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 19.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baümler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 20.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 21.Porwollik, S., R. M.-Y. Wong, S. H. Sims, R. M. Schaaper, D. M. DeMarini, and M. McClelland. 2001. The ΔuvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat. Res. 483:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Belkum, A., M. Struelens, A. De Visser, H. Verbrugh, and M. Tybayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westin, L., C. Miller, D. Vollmer, D. Canter, R. Radtkey, M. Nerenberg, and J. P. O'Connell. 2001. Antimicrobial resistance and bacterial identification utilizing microelectronic chip array. J. Clin. Microbiol. 39:1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]