Abstract
The purpose of this study was to determine the bacterial diversity in advanced noma lesions using culture-independent molecular methods. 16S ribosomal DNA bacterial genes from DNA isolated from advanced noma lesions of four Nigerian children were PCR amplified with universally conserved primers and spirochetal selective primers and cloned into Escherichia coli. Partial 16S rRNA sequences of approximately 500 bases from 212 cloned inserts were used initially to determine species identity or closest relatives by comparison with sequences of known species or phylotypes. Nearly complete sequences of approximately 1,500 bases were obtained for most of the potentially novel species. A total of 67 bacterial species or phylotypes were detected, 25 of which have not yet been grown in vitro. Nineteen of the species or phylotypes, including Propionibacterium acnes, Staphylococcus spp., and the opportunistic pathogens Stenotrophomonas maltophilia and Ochrobactrum anthropi were detected in more than one subject. Other known species that were detected included Achromobacter spp., Afipia spp., Brevundimonas diminuta, Capnocytophaga spp., Cardiobacterium sp., Eikenella corrodens, Fusobacterium spp., Gemella haemoylsans, and Neisseria spp. Phylotypes that were unique to noma infections included those in the genera Eubacterium, Flavobacterium, Kocuria, Microbacterium, and Porphyromonas and the related Streptococcus salivarius and genera Sphingomonas and Treponema. Since advanced noma lesions are infections open to the environment, it was not surprising to detect species not commonly associated with the oral cavity, e.g., from soil. Several species previously implicated as putative pathogens of noma, such as spirochetes and Fusobacterium spp., were detected in at least one subject. However, due to the limited number of available noma subjects, it was not possible at this time to associate specific species with the disease.
Noma, or cancrum oris, is a severe gangrenous disease of the oral cavity that generally affects children in developing countries and is characterized by a strong putrid odor and a rapid (within days) necrotizing destruction of soft and hard tissue, including bone (6, 8, 9, 14). Before the introduction of antibiotics, such as metronidazole (16), noma was often fatal, with as high as 75% mortality. The risk factors for noma are chronic malnutrition, dehydration, poor oral hygiene, and a state of debilitation resulting from human immunodeficiency virus infection, measles, and other childhood diseases that are prevalent in the tropics (9). The disease begins with the ulceration and necrosis of the interdental papillae and extends into the surrounding tissue, causing bone exposure, bone sequestration, and penetration into adjacent cheek mucosa (6). Consequently, children with advanced lesions quite literally have gaping holes in their cheeks. Survivors have severe problems with eating, drinking, and speaking and many die of complications of the infection or by starvation because they are unable to chew due to the facial mutilation (1). Although noma has essentially disappeared in affluent countries, the disease is still reported in many countries of Africa, the Dominican Republic, Pakistan, Paraguay, Peru, Uruguay, and India. A wide range of incidence has been reported at about 1 in 1,000 children in Nigeria and Senegal (1, 9). However, in the most-affected communities, there are as many as 12 cases of noma per 1,000 children. Worldwide, the World Health Organization estimates that the number of children less than 6 years old who contract noma is about 200,000 per year (1).
The bacterial etiology of noma has not been firmly established. It has been suggested that noma results from a mixed infection of oral and extraoral opportunistic pathogens (9, 10, 11). Spirochetes and species of Fusobacterium have long been suggested to play a role in the disease process (10, 13, 33). Based on early ultrastructural studies, a long filamentous, gram-positive rod and spirochetes were implicated as potential etiologic agents of noma (24). More recently, it has been proposed that at least certain cases of noma result from a zoonotic infection with Fusobacterium necrophorum (10, 11), an etiologic agent of foot rot in sheep and other livestock.
Using culture-independent molecular methods, it has been determined that there are 500 to 600 bacterial species present in the oral cavity and that about 40 to 50% of these species are currently unrecognized or have not yet been cultivated (26). The purpose of this study was to use similar methods to determine the prevalent species and phylotypes in advanced lesions of children with noma. A secondary purpose was to compare the bacterial composition and diversity of advanced noma lesions with those of other oral sites, such as subgingival plaque of different periodontal disease states and periodontal health, supragingival plaque on tooth surfaces, and epithelial cells from the subgingival crevice or tongue dorsum.
MATERIALS AND METHODS
Subjects.
Noma subjects used in this study have been previously described in detail (10). All subjects were from northwestern Nigeria within a 2 h drive to Sokoto City. The four subjects that were analyzed were 5 to 15 years old and were malnourished. The children presented with lesions that had been present from 6 weeks to 2 years. Clinical characterization of each subject is as follows.
(i) Subject SK2.
Subject SK2 was a 5-year-old female who weighed 10 kg and was 80 cm tall. She had a 5-mm-diameter hole from the right alar of the nose to a point 3 mm superior to the right angle of the mouth. The alveolar bone of the alveolar ridge and palate was destroyed. The inner concha of the right side of nose was also exposed.
(ii) Subject SK5.
Subject SK5 was a 7-year-old female who weighed 12 kg and was 90 cm tall. She had severe destruction of the lip around the left angle of the mouth and nose. There was severe fibrosis and ankylosing of the right angle of the mouth.
(iii) Subject SK7.
Subject SK7 was a 15-year-old female who weighed 41 kg and was 1.58 m tall. She had destruction of the left cheek and alar of the left side of the nose that left a space about 5 mm in diameter. A strand of the vermilion border tissue was still visible. A shiny fibrosis of the skin around the lesion and left alveolar ridge was also destroyed.
(iv) Subject SK10.
Subject SK10 was a 15-year-old male who weighed 28 kg and was 1.4 m tall. He had destruction of the upper left lip, alveolar bone and palate, and lower left lip, which was moderately ankylosed. In addition, he had drooping of the left eye.
Microbiological sampling.
Oral sites were isolated with cotton rolls to prevent salivary contamination. Sterile endodontic paper points were intraorally inserted directly into the sites of advanced noma lesions, either between the tooth surface and gingival tissue or at the advancing margin of tissue damage (10). Samples were directly suspended in 50 μl of solution containing 50 mM Tris buffer, pH 7.6; 1 mM EDTA, pH 8.0; and 0.5% Tween 20. NaOH was added at a final concentration of 0.5 N to inactivate the sample and to preserve the DNA. Samples were stored at room temperature for approximately 2 weeks until shipped to the Paster laboratory, where they were stored at −20°C until processed. Samples were pH adjusted to about 7.5, and proteinase K (200 μg/ml) was added. The samples were then heated at 55°C for 2 h. Proteinase K was inactivated by heating at 95°C for 5 min.
Amplification of 16S rRNA cistrons by PCR and purification of PCR products.
The 16S rRNA genes were amplified under standard conditions using two different primer sets—universally conserved and spirochete selective. The sequences of the primers have been previously reported (26). Results obtained using the spirochete-selective primers indicate that 85% of the clones have spirochetal inserts (7). PCR was performed in thin-walled tubes with a Perkin-Elmer 9700 Thermocycler. One microliter of the DNA template was added to a reaction mixture (50-μl final volume) containing 20 pmol of each primer, 40 nmol of each deoxynucleoside triphosphate, and 1 U of Taq 2000 polymerase (Stratagene, La Jolla, Calif.) in buffer containing Taqstart Antibody (Sigma Chemical Co.). In a hot-start protocol, samples were preheated at 95°C for 8 min, and this was followed by amplification using the following conditions: denaturation at 95°C for 45 s, annealing at 60°C for 45 s, and elongation for 1.5 min, with an additional 5 s for each cycle. A total of 30 cycles were performed and then followed by a final elongation step at 72°C for 10 min. The results of PCR amplification were examined by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light.
Cloning procedures.
Prior to cloning, the PCR-amplified 16S ribosomal DNA (rDNA) fragments were purified and concentrated using Microcon 100 (Amicon), followed by the QIAquick PCR purification kit (QIAGEN). The purified PCR product was then blunted with the Pfu polymerase in preparation for ligation. Using the Zero Blunt PCR Cloning Kit (Invitrogen), the blunted product was ligated into the pCR-Blunt vector. Transformation was done using competent Escherichia coli TOP10 cells provided by the manufacturer. The transformed cells were then plated onto Luria-Bertani agar plates supplemented with kanamycin and incubated overnight at 37°C. Colonies were then placed into 40 μl of 10 mM Tris. One microliter of this suspension with the M13 (−40) forward primer and the M13 reverse primer were used in standard PCRs to identify clones with the correct size of insert.
16S rRNA sequencing.
Purified DNA from PCR was sequenced using an ABI prism cycle-sequencing kit (BigDye Terminator Cycle Sequencing kit with AmpliTaq DNA Polymerase FS; Perkin-Elmer). The primers used for sequencing have been previously described (26). Quarter dye chemistry was used with 80 μM concentrations of primers and 1.5 μl of PCR product in a final volume of 20 μl. Cycle sequencing was performed using an ABI 9700 Thermocycler with 25 cycles of denaturation at 96°C for 10 s and annealing and extension at 60°C for 4 min. Sequencing reactions were run on an ABI 377 DNA sequencer.
16S rRNA sequencing and data analysis of unrecognized inserts.
A total of 212 clones with the correct size of insert, approximately 1500 bases, were analyzed. In these studies, approximately 500 bases were obtained first to determine identity or approximate phylogenetic position. Full sequences (about 1,500 bases using five to six additional sequencing primers [26]) were obtained for most of the novel phylotypes and used for phylogenetic analysis. For identification of the closest relatives, sequences of the unrecognized inserts were compared to the 16S rRNA gene sequences of more than 4,000 microorganisms in our database and of the 16,000 sequences in the Ribosomal Database Project (23) and GenBank. Programs for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and phylogenetic tree construction were written by F. E. Dewhirst (27). The similarity matrices were corrected for multiple base changes at single positions by the method of Jukes and Cantor (15). Similarity matrices were constructed from the aligned sequences by using only those sequence positions at which 90% of the strains had data. Phylogenetic trees were constructed using the neighbor-joining method of Saitou and Nei (29). TREECON, a software package for the Microsoft Windows environment, was used for the construction and drawing of evolutionary trees (34). Two hundred bootstrap trees were generated, and bootstrap confidence levels were determined using the TREECON program.
We are aware of the potential creation of 16S rDNA chimera molecules assembled during the PCR (19). Percentages of chimeric inserts in 16S rRNA libraries ranged from 1 to 15%. Chimeric sequences were identified by using the Chimera check program of the Ribosomal Database Project package, by tree analysis, or by base signature analysis.
Nucleotide sequence accession numbers.
The complete 16S rRNA gene sequences of clones representing novel phylotypes defined in this study, sequences of known species not previously reported, and published sequences are available for electronic retrieval from GenBank, EMBL, and DDBJ nucleotide sequence databases under the accession numbers shown in Fig. 1 . The newly assigned accession numbers for the novel phylotypes are as follows: for clone AW149, AF393477; for clone BF017, AF393478; for clone AV069, AF385529; for clone AW030, AF385533; for clone AV011a, AF385528; for clone CA004, AF385538; for clone AY088, AF385546; for clone AZ002, AF385547; for clone CA002, AF385537; for clone CA006, AF385539; for clone AV085, AF385530; for clone AW086, AF385535; for clone BZ008, AF393479; for clone CA007, AF385540; for clone CB003, AF385541; for clone AW006, AF385532; for clone AV005b, AF385527; for clone BZ013, AF385536; for clone AV100, AF385531; for clone AY017, AF385544; and for clone AW032, AF393476.
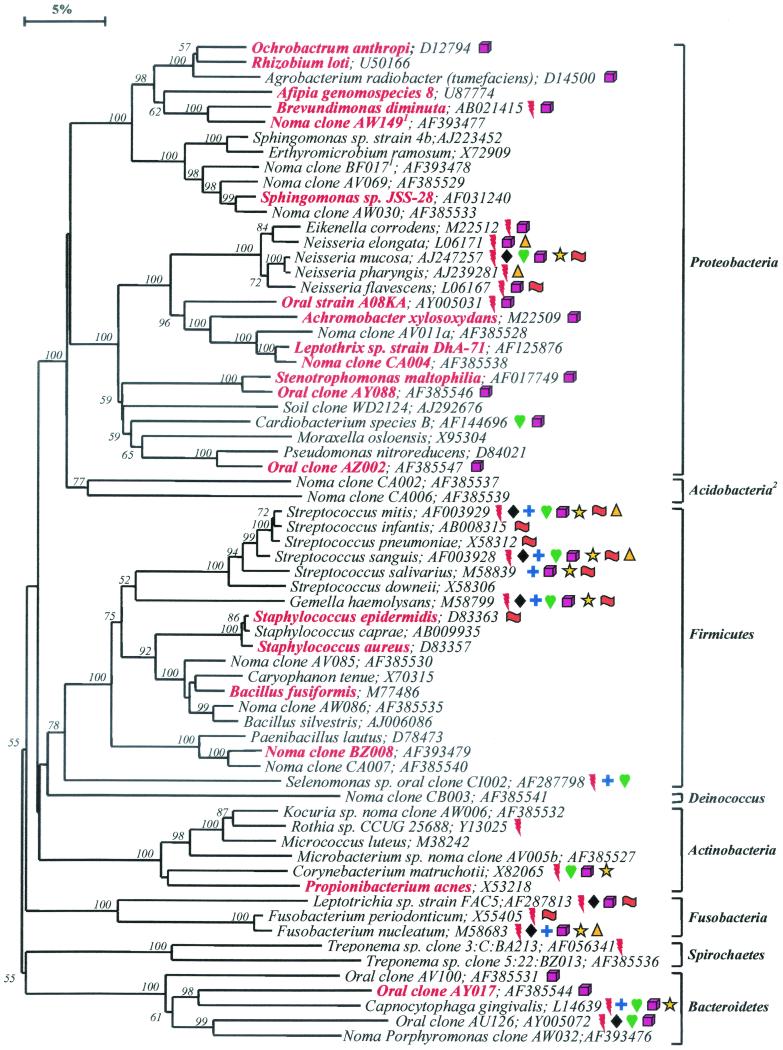
FIG. 1.
Phylogenetic tree of full 16S rDNA sequences of clonal libraries derived from advanced noma lesions. Only those species or phylotypes detected in noma lesions are shown. The information presented includes bacterial species or phylotype and GenBank accession number. Species or phylotypes detected in more than one subject are highlighted in red boldface lettering and are also listed in Table 1. Color-coordinated symbols are used to identify those species or phylotypes that were also detected in oral sites other than noma lesions, e.g., in subgingival plaque of healthy and diseased sites (26), in supragingival plaque on tooth surfaces (2), on or in subgingival crevicular epithelial cells (5), and on epithelial cells of the tongue dorsum (18). The superscripted “1” indicates a partial sequence of about 500 bp. The bar at top represents a 5% difference in nucleotide sequences. The superscripted “2” indicates a newly described bacterial phylum. Two hundred bootstrap trees were generated, and bootstrap confidence levels as percentages (only values over 50%) are shown at tree nodes. Symbols: red lightning bolt, refractory periodontitis subgingival plaque; black diamond, periodontitis subgingival plaque; blue cross, acute necrotizing ulcerative gingivitis subgingival plaque; golden triangle, necrotizing ulcerative periodontitis subgingival plaque; green heart, healthy subgingival plaque; yellow star, dental supragingival plaque; fuchsia cube, on or in crevicular epithelial cells; orange flag, on or in tongue dorsum epithelial cells.
RESULTS AND DISCUSSION
Partial sequences of about 500 bp were obtained for 212 16S rRNA clones in order to identify the prevalent bacterial species present in advanced noma lesions. Nearly complete sequences of approximately 1,500 bp were obtained for most of the potentially novel species, or phylotypes. The bacterial diversity in these lesions was impressive—67 bacterial species or phylotypes from eight bacterial phyla were detected (Fig. 1). Fifty-two (63%) were known cultivable species or strains within known genera, and 25 (37%) were phylotypes, i.e., species that have not yet been cultivated or are currently unrecognized. Sixteen of these phylotypes (designated as “noma clones” in Fig. 1) were novel in that they have not been detected, thus far, in any oral site other than in noma lesions. “Phylotype” was defined as a cluster of clone sequences that differed from known species by at least 30 bases (or 2% difference in full-sequence comparisons) and was at least 99% similar to other members of its cluster.
In subgingival plaque from healthy subjects and subjects with various forms of periodontal disease, we previously detected 347 species or phylotypes, which fell into nine different bacterial phyla (26). Representatives of two additional bacterial phyla, Deinococcus and Acidobacteria, were detected in noma libraries (Fig. 1). Deinococcus is a phylum comprised of radioresistant bacterial species. Acidobacteria is a newly described phylum that contains only a few cultivable species, such as those in the genera Acidobacterium and Holophaga, but mostly uncultivated phylotypes from soil, sediment, and activated sludge (21). Overall, bacteria detected from the oral cavity now fall into 11 different bacterial phyla that comprised at least 600 different species or phylotypes (26).
Figure 1 depicts the breadth of bacterial diversity in advanced noma lesions—only those species and phylotypes detected in these lesions are shown. Color-coordinated symbols are used to identify those species or phylotypes that were also detected in subgingival plaque of healthy subjects and subjects with various levels of periodontal disease (26, 28), in supragingival plaque on tooth surfaces of healthy children and children with rampant caries (2), on epithelial cells of the tongue dorsum of healthy subjects and subjects with halitosis (5), and on or in subgingival crevicular epithelial cells from healthy and diseased subjects (18).
Nineteen of the species or phylotypes were detected in more than one subject (Table 1 and highlighted in boldface red lettering in Fig. 1). Fifteen of these species or phylotypes were common to subject SK5 and subject SK7 (Table 1). Although the total number of clones analyzed from subjects SK2 and SK10 was limited, there was, nonetheless, some overlap in bacterial distribution among subjects (Table 1). These commonly detected species include Achromobacter (Alcaligenes) xylosoxidans, Afipia spp., Brevundimonas diminuta, Ochrobactrum anthropi, Propionibacterium acnes, Staphylococcus spp., and Stenotrophomonas maltophilia. S. maltophilia was detected in three of the subjects and represented approximately one-third of the clones in one subject (Table 1). Some of these species have been reported as putative pathogens in extraoral infections. For example, A. xylosoxidans and S. maltophilia are considered to be emerging pathogens in pulmonary infections such as cystic fibrosis and pneumonia, endocarditis, meningitis, and other nosocomial infections (4). O. anthropi is another opportunistic pathogen involved in nosocomial infections, such as catheter-related sepsis or infectious endocarditis (22). A common and compounding feature of these organisms is that they are naturally resistant to multiple antibiotics (12).
TABLE 1.
Distribution of prevalent species or phylotypes in noma subjectsa
| Species or phylotype | No. of clones detected in subject:
|
|||
|---|---|---|---|---|
| SK5 (n = 94) | SK7 (n = 90) | SK2 (n = 19) | SK10 (n = 9) | |
| Achrombacter xylosoxidans | 9 | 1 | ||
| Afipia genomospecies 8 | 2 | 2 | ||
| Bacillus fusiformis | 2 | 3 | ||
| Brevundimonas diminuta | 1 | 6 | ||
| Leptothrix strain DhA-71 | 1 | 1 | ||
| Noma clone AW149 | 1 | 1 | ||
| Noma clone BZ008 | 1 | 1 | ||
| Noma clone CA004 | 1 | 1 | ||
| Ochrobactrum anthropi | 11 | 5 | ||
| Oral clone AY017 | 1 | 1 | ||
| Oral clone AY088 | ||||
| Oral clone AZ002 | 2 | 1 | ||
| Oral strain A08KA | 14 | 1 | ||
| Propionibacterium acnes | 1 | 1 | ||
| Rhizobium loti | 1 | 1 | ||
| Sphingomonas strain JSS-28 | 1 | 3 | ||
| Staphylococcus aureus | 2 | 2 | ||
| Staphylococcus epidermidis | 1 | 1 | ||
| Stenotrophomonas maltophilia | 29 | 1 | 1 | |
Prevalent species that are highlighted in red in Fig. 1.
Additional known oral species that were detected in at least one of the subjects include Capnocytophaga spp., Cardiobacterium sp., Eikenella corrodens, Fusobacterium spp., Gemella haemolysans, Neisseria spp., and Streptococcus spp. Many of these species have been associated with periodontal diseases (30), and some of the species, such as G. haemolysans, Cardiobacterium spp., and E. corrodens, are opportunistic pathogens that have been implicated in infectious endocarditis (3, 17).
In general, the microbiota of noma lesions was comprised of species not commonly associated with periodontal disease or health. Since only limited information is available on the identification of specific species associated with noma lesions, it is possible that the relatively atypical microbiota detected may be attributed to the infection itself. Clearly, the microbial analysis of noma lesions of additional subjects must be performed before definitive conclusions can be made. Furthermore, since these advanced noma lesions were infections open to the environment as well as under unsanitary conditions, it was not surprising to also detect species not commonly associated with the oral cavity. Species of Flavobacterium, Microbacterium, Sphingomonas, Bacillus, Paenibacillus, and Rhizobium (Fig. 1) are widely distributed in the environment, e.g., soil, and rarely cause infections in humans.
Fusobacterium spp. have long been associated with noma infections (10, 13, 32, 33). Recently, F. necrophorum, an opportunistic pathogen that causes numerous necrotic conditions (necrobacillosis) such as bovine hepatic abscesses, ruminant foot abscesses, and human oral infections (31), was recovered from seven of eight advanced noma lesions (10). Since the infected children were in close contact with livestock (i.e., they eat and sleep with their animals), the authors concluded that noma may be a zoonotic infection caused by F. necrophorum (10, 11). Since F. necrophorum was isolated from these samples on selective media, it probably represents less than 1% of the total viable population, which is below the level of detection for the methods used in this study. There was a notable absence of other species that one might expect to see in these infections, such as Actinomyces spp., Streptococcus intermedius, and those species commonly associated with periodontal disease and health. However, in our ongoing studies of determining the 16S rRNA genes sequences of all oral bacteria, our results demonstrated that our procedures do indeed recover a broad spectrum of bacterial species (26). Note that these species may still be present but below the limit of detection.
Spirochetes have also been associated with noma infections (20, 24, 33), but typically are observed at lower levels compared to other organisms. Spirochetes were observed in some of the samples (10). Indeed in one of the samples, we were able to detect two uncultivated phylotypes of treponemes, one of which (clone BZ013) was unique to noma infections (Fig. 1).
These molecular studies are but the first step in understanding the bacterial etiology of noma—to identify those species, both cultivable and not yet cultivated, that are associated with advanced noma lesions. Once these organisms are identified, 16S rDNA-based probes can be used in larger clinical studies using DNA-DNA hybridization techniques (2, 25) to compare the population distribution of specific species found in samples from noma subjects with the distribution in clinically healthy subjects. However, in subsequent studies, we will focus on early noma lesions rather than gangrenous lesions, since many organisms in these advanced infections have had the opportunity to proliferate and potentially mask the presence of putative etiologic agents.
While the present investigation is heavily weighted toward examining the microbial side of noma, we are fully cognizant of the potential for host factors playing a crucial role in this disease. These studies may provide insight for the identity of putative pathogens in other oral diseases, such as periodontitis, acute necrotizing ulcerative gingivitis and refractory periodontitis. In other words, specific bacterial species that can wreak havoc in a severely compromised host may serve as putative pathogens (or at least predictive microorganisms) in the more-common oral diseases.
Acknowledgments
This work was supported by Public Health Service grants DE11443 and DE10374 to B.J.P. and F.E.D., Public Health Service grant 3043TW0907, and a grant to C.O.E. and W.A.F. from the Nestle Foundation, Lausanne, Switzerland.
REFERENCES
- 1.Barmes, D. E., C. O. Enwonwu, M.-H. Leclercq, D. Bourgeois, and W. A. Falkler. 1997. The need for action against oro-facial gangrene (noma). Trop. Med. Int. Health 2:1111-1114. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berbari, E. F., F. R. Cockerill III, and J. M. Steckelberg. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72:532-542. [DOI] [PubMed] [Google Scholar]
- 4.Beringer, P. M., and M. D. Appleman. 2000. Unusual respiratory bacterial flora in cystic fibrosis: microbiologic and clinical features. Curr. Opin. Pulm. Med. 6:545-550. [DOI] [PubMed] [Google Scholar]
- 5.Boches, S. K., P. M. Mitchell, J. L. Galvin, W. J. Loesche, C. E. Kazor, F. E. Dewhirst, and B. J. Paster. 2000. Cultivable and uncultivable bacteria on the healthy tongue dorsum. J. Dent. Res. 79:396. [Google Scholar]
- 6.Deeb, G. R., W. Y. Yih, R. G. Merrill, and R. C. Lundeen. 1999. Noma: report of a case resulting in bony ankylosis of the maxilla and mandible. Dentomaxillofac. Radiol. 28:378-382. [DOI] [PubMed] [Google Scholar]
- 7.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 8.Enwonwu, C. O. 1995. Noma: a neglected scourge of children in sub-Saharan Africa. Bull. W. H. O. 73:541-545. [PMC free article] [PubMed] [Google Scholar]
- 9.Enwonwu, C. O., W. A. Falkler, Jr., E. O. Idigbe, B. M. Afalabi, M. Ibrahim, D. Onwujekwe, O. Savage, and B. I. Meeks. 1999. Pathogenesis of cancrum oris (noma): confounding interactions of malnutrition with infection. Am. J. Trop. Med. Hyg. 60:223-232. [DOI] [PubMed] [Google Scholar]
- 10.Falkler, W. A., Jr., C. O. Enwonwu, and E. O. Idigbe. 1999a. Isolation of Fusobacterium necrophorum from cancrum oris (noma). Am. J. Trop. Med. Hyg. 60:150-156. [DOI] [PubMed]
- 11.Falkler, W. A., Jr., C. O. Enwonwu, and E. O. Idigbe. 1999b. Microbiological understandings and mysteries of noma (cancrum oris). Oral Dis. 5:150-155. [DOI] [PubMed]
- 12.Higgins, C. S., S. M. Murtough, E. Williamson, S. J. Hiom, D. J. Payne, A. D. Russell, and T. R. Walsh. 2001. Resistance to antibiotics and biocides among non-fermenting Gram-negative bacteria. Clin. Microbiol. Infect. 7:308-315. [DOI] [PubMed] [Google Scholar]
- 13.Horning, G. M. 1996. Necrotizing gingivostomatitis: NUG to noma. Compendium 17:951-962. [PubMed] [Google Scholar]
- 14.Idigbe, E. O., C. O. Enwonwu, W. A. Falkler, M. M. Ibrahim, D. Onwujekwe, B. M. Afolabi, K. O. Savage, and V. I. Meeks. 1999. Living conditions of children at risk for noma: Nigerian experience. Oral Dis. 5:156-162. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 16.Lacour, M., and M. Castets. 1968. Sur un cas de noma traité par le métronidazole. Bull. Soc. Med. Afr. Noire 3:701-703. [PubMed] [Google Scholar]
- 17.La Scola, B., and D. Raoult. 1998. Molecular identification of Gemella species from three patients with endocarditis. J. Clin. Microbiol. 36:866-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, A. M., P. M. Mitchell, L. N. Stokes, C. E. Kazor, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2002. Comparative molecular analysis of tongue bacteria in halitosis and in health. J. Dent. Res. 81:A-364. [Google Scholar]
- 19.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 20.Loesche, W. J. 1976. Periodontal disease and the treponemes, p. 261-275. In W. J. Loesche (ed.), The biology of parasitic spirochetes. Academic Press, New York, N.Y.
- 21.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 22.Mahmood, M. S., A. R. Sarwari, M. A. Khan, Z. Sophie, E. Khan, and S. Sami. 2000. Infective endocarditis and septic embolization with Ochrobactrum anthropi: case report and review of literature. J. Infect. 40:287-290. [DOI] [PubMed] [Google Scholar]
- 23.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrell, B. R., S. W. Joesph, L. J. Casazza, and J. F. Duncan. 1981. Bacterial bone resorption in noma (gangrenous stomatitis). J. Oral Path. 10:173-185. [DOI] [PubMed] [Google Scholar]
- 25.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S rRNA sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 28.Russell, M. K., T. Alpagot, S. K. Boches, J. L. Galvin, F. E. Dewhirst, and B. J. Paster. 2001. Bacterial species and phylotypes in necrotizing ulcerative periodontitis. J. Dent. Res. 80:167.. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 31.Tan, Z. L., T. G. Nagaraja, and M. M. Chengappa. 1996. Fusobacterium necrophorum infections: virulence factors, pathogenic mechanism and control measures. Vet. Res. Commun. 20:113-140. [DOI] [PubMed] [Google Scholar]
- 32.Tempest, M. N. 1966. Cancrum oris. Br. J. Surg. 53:949-969. [DOI] [PubMed] [Google Scholar]
- 33.Uohara, G. I., and M. J. Knapp. 1967. Oral fusospirochetosis and associated lesions. Oral Surg. Oral Med. Oral Pathol. 24:113-123. [DOI] [PubMed] [Google Scholar]
- 34.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Applic. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]