Abstract
Escherichia coli O15:K52:H1 is a significant extraintestinal pathogen in Europe (G. Prats et al., J. Clin. Microbiol. 38:201-209, 2000). To search for evidence of this clonal group outside of Europe, 75 non-European E. coli isolates of serogroup O15 were compared with five members of the O15:K52:H1 clonal group from Barcelona, Spain, according to genomic background, virulence genotypes, and antimicrobial resistance profiles. Amplification phylotyping showed that 16 (21%) of the 75 non-European O15 isolates corresponded with the O15:K52:H1 clonal group. The 16 non-European O15:K52:H1 clonal group members represented diverse geographic locales. They were isolated almost exclusively from humans with extraintestinal infections and accounted for 50% of all O15 isolates from five human clinical collections studied. Most non-European clonal group members exhibited a consensus virulence factor profile that included the F16 or F7-2 papA alleles (P fimbrial structural subunit), papG allele II (P fimbrial adhesin), iha (putative adhesin siderophore), and iutA (aerobactin receptor). This resembles the virulence profiles of (i) European representatives of the O15:K52:H1 clonal group and (ii) phylogenetically related “clonal group A,” a recently recognized significant contributor to trimethoprim-sulfamethoxazole resistance in the United States (A. R. Manges et al., N. Engl. J. Med. 345:1007-1013, 2001). Antimicrobial resistance profiles were variable, and resistance was inconsistently transferred by conjugation. These findings indicate that the O15:K52:H1 clonal group is broadly distributed beyond Europe, exhibits previously unrecognized phenotypic and genotypic diversity, and contributes significantly to extraintestinal infections in humans.
Most extraintestinal infections due to Escherichia coli, including urinary tract infections (UTIs), sepsis, and neonatal meningitis, are caused by distinctive “virulent clones” of E. coli. Such virulent clones exhibit diverse specialized virulence factors (VFs) that enable the organisms to overcome or subvert host defenses, injure or invade host cells, and incite a noxious inflammatory host response, thereby producing disease. Virulent clones can be recognized by their characteristic O:K:H serotypes or by genetic typing methods. They are globally distributed and usually cause sporadic infections; however, nosocomial and community source outbreaks due to these organisms have been reported (27, 42).
E. coli serotype O15:K52:H1 does not represent one of the traditionally recognized “virulent clones” of extraintestinal pathogenic E. coli (34, 35). Members of this serotype first came to attention as significant extraintestinal pathogens when they caused a large-scale epidemic of UTI, septicemia, and diverse other extraintestinal infections in South London, England, in the winter of 1986 to 1987, in association with a distinctive multiple antimicrobial resistance profile (32, 36; D. S. Reeves, R. A. Bailey, M. J. Bywater, H. A. Holt, A. P. MacGowan, and R. J. Marshall, Letter, Lancet i:929, 1987; D. J. Waghorn, D. J., T. W. J. Kelly, and W. Gibbins, Letter, J. Hosp. Infect. 11:192-193, 1987; E. D. Wright and R. M. Perinpanayagam, Letter, Lancet i:556-557, 1987). This serotype later was identified as a prominent cause of E. coli bacteremia in Copenhagen, Denmark (33), and Terrassa, Spain (D. Dalmau, F. Navarro, B. Mierlis, J. Blanco, J. Garau, and G. Prats, Letter, J. Hosp. Infect. 34:233-234, 1996).
Recently, in a prospective study from Barcelona, Spain, E. coli O15:K52:H1 was shown to be a significant cause of community-acquired and nosocomial UTI (38). Compared with E. coli urine isolates of other serotypes, those of serotype O15:K52:H1 were more likely to cause clinical pyelonephritis and to infect young adults, evidence of enhanced virulence. O15:K52:H1 strains from England and various centers in Spain were shown to be clonally derived and to exhibit a homogeneous VF profile, including the presence of pap (pilus associated with pyelonephritis), papG allele II (P fimbrial adhesin), the F16 papA allele (P fimbrial structural subunit), and iutA (aerobactin receptor), and the absence of sfa, hly, and cnf (S fimbriae, hemolysin, and cytotoxic necrotizing factor) but to have diverse antimicrobial susceptibility patterns (38). Randomly amplified polymorphic DNA (RAPD) analysis showed that this clonal group derives from virulence-associated phylogenetic group D (38), as defined within the E. coli Reference (ECOR) collection by multilocus enzyme electrophoresis or sequence typing (3, 9, 25). This clonal group also was recently shown to be closely related phylogenetically to “clonal group A,” a newly emerged clonal group of uropathogenic E. coli that is broadly disseminated in the United States and contributes substantially to the current epidemic of trimethoprim-sulfamethoxazole resistance in uncomplicated cystitis and pyelonephritis (20, 27).
To date, the O15:K52:H1 clone has been described only from Western Europe (38). The occurrence of two strains of this O:K:H serotype among urosepsis isolates from Seattle, Wash., that exhibited a VF profile consistent with that of the European members of this clone (23), and the prominence of serogroup O15 among E. coli isolates from pregnant women with pyelonephritis in Galveston, Tex. (K. Nguyen, A. Hart, and B. J. Nowicki, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. B-177, p. 85, 1998), prompted us to search more broadly for this clonal group. Specifically, we sought to define the prevalence, geographic extent, host range, clinical syndrome capabilities, VF profiles, and antimicrobial resistance patterns of the E. coli O15:K52:H1 clonal group outside of Europe.
(These findings were presented in part at the 99th General Meeting of the American Society for Microbiology, May 30 to June 3 1999, Chicago, Ill. [abstr. B/D-11].)
MATERIALS AND METHODS
Strains.
Seventy-five non-European E. coli isolates of serogroup O15 from diverse sources were compared with five UTI isolate representatives of the E. coli O15:K52:H1 clone from Barcelona, Spain, which served as the reference set (38). The 75 non-European O15 isolates were from various locations in the United States (n = 68), Canada (n = 2), or elsewhere in the world (n = 5). Known host species included human (n = 39), turkey (n = 5), dog (n = 2), cattle (n = 11), pig (n = 3), goose (n = 1), horse (n = 2), mink (n = 1), chicken (n = 8), deer (n = 1), and rabbit (n = 2) sources. Five strains were water isolates. Twenty-six of the human isolates represented all available serogroup O15 isolates from five serotyped human clinical collections of extraintestinal E. coli (15, 23, 29; A. Kaul, unpublished data; Nguyen et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998). The clinical syndromes encompassed by these five collections (inclusive dates; proportion from serogroup O15) included urosepsis in adults (1984 to 1987; 3/68) (23), gestational pyelonephritis (1994 to 1997; 10/81) (Nguyen et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998), cystitis in children (1987 to 1990; 6/38) (15), diverse-source bacteremia in adults (1988 to 1991; 4/187) (11, 29), and symptomatic or asymptomatic bacteriuria in women (1997 to 1998; 3/94) (Kaul, unpublished). The remaining 49 predominantly animal isolates represented all additional serogroup O15 isolates that were available from the Escherichia coli Reference Center (ECRC, University Park, Pa.). These isolates were received by the ECRC from 1984 through 1998, as indicated by the first two digits of their six-digit strain designations (Fig. 1 ), but their dates of primary isolation were not available.
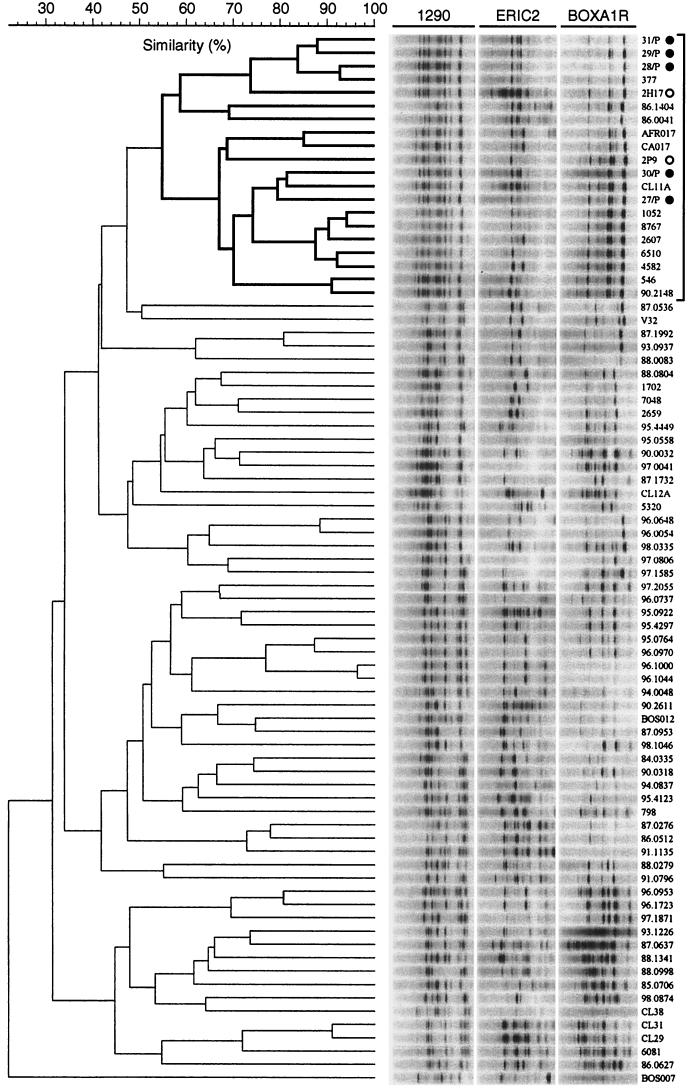
FIG.1.
Phylogenetic relationships among E. coli isolates of serogroup O15. The dendrogram was inferred according to UPGMA based on composite genomic profiles as generated by PCR with RAPD primer 1290 (left) and rep-PCR primers ERIC2 (center) and BOXA1R (right). Strain designations are shown on the far right. The subcluster corresponding to the O15:K52:H1 clonal group is demarcated in boldface lines at the top of the dendrogram. Bullets indicate the five Spanish O15:K52:H1 control strains; open circles indicate the two previously known O15:K52:H1 urosepsis isolates from Seattle. Gel strips as shown are schematic digital reconstructions and hence underestimate the resolution of the actual gel images. Bacteremia isolate CA059 (not included in Fig. 1) also was indistinguishable from the Spanish O15:K52:H1 strains according to composite amplification phylotyping.
Serotyping.
O:K:H serotypes for the O15:K52:H1 isolates from Barcelona (n = 5) and Seattle (n = 2) were as previously reported (21, 38). O:H serotypes or O antigens for the remaining isolates determined by the ECRC. Confirmatory O:K:H serotypes were newly determined for selected strains by the International Escherichia and Klebsiella Centre (World Health Organisation, Copenhagen, Denmark).
PCR genomic patterns and clonal analysis.
Composite amplification profiling provides a convenient surrogate for reference phylotyping methods, such as multilocus enzyme electrophoresis, to assess phylogenetic relationships among E. coli isolates (5, 17, 44) (Fig. 1). Consequently, genomic profiles were generated by PCR for all strains by using (separately) three different primers (14, 17, 19). RAPD analysis was done by using as a primer decamer oligonucleotide 1290 (1). Repetitive element (rep-) PCR was done by using the enterobacterial repetitive intergenic consensus (ERIC) primer ERIC2 and primer BOXA1R (43), as previously described (17). Boiled lysates were used as the amplification template. Amplification conditions were as previously described (43), except for the substitution of Ready-to-Go PCR beads (Pharmacia, Piscataway, N.J.). Banding patterns in ethidium bromide-stained agarose gels were digitally analyzed by using a computerized image capture and analysis system (Gel Doc and Molecular Analyst; Bio-Rad, Hercules, Calif.) (14, 19). Amplification patterns from the three separate primers were digitally combined head-to-tail to create a single “virtual” composite pattern for each strain (12, 14, 17, 19). Pearson's correlation coefficient was used to derive similarity values for all pairwise comparisons between composite genomic patterns for the 80 isolates, based on analog densitometric scans of gel tracks (12, 14, 17, 19). Dendrograms were inferred from the resulting similarity matrices by using the unweighted pair group method with arithmetic averages (UPGMA) (41). Of the several dendrograms that were constructed for the same image data set by using different settings within Molecular Analysis (not shown), the one which provided the best resolution of the five Spanish O15:K52:H1 control strains was used to define the O15:K52:H1 clonal group (Fig. 1). In this dendrogram, all strains included within the smallest cluster that contained the five Spanish controls were defined as being members of the O15:K52:H1 clonal group.
Multiplex PCR assays for virulence genes and papA alleles.
Thirty-one genes encoding putative VFs of extraintestinal pathogenic E. coli were detected by using a multiplex PCR assay, as previously described (Table 1) (22, 23). The group II capsule synthesis region kpsMT II was detected also by probe hybridization in dot blots, as previously described, to identify the K2 kpsMT II variant (23). Positive and negative control strains were as previously described (22, 23). All strains were tested for the 12 known alleles of papA (which encodes PapA, the major structural subunit and major antigenic determinate of P fimbriae), including alleles F7-1, F7-2, F8-F16, and F48, by using an allele-specific multiplex PCR assay, as previously described (24). All assays were done in duplicate by using independently prepared template DNA samples, with any discrepancies investigated further as needed.
TABLE 1.
Epidemiological and bacteriological characteristics of 80 E. coli isolates of serogroup O15a
| Strain | Serotype | Source | Syndrome | Location | F type |
pap
|
papG allele | sfa/foc | focG | afa/dra | bmaE | afD | iha | flmH |
kpsMT
|
fyuA | IroN | IutA | hlyA | cnf1 | cdtB | beA | traT | cvaC | malX | Antibiotics | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | EF | G | II | III | K1 | |||||||||||||||||||||||||
| O15:K52:H1 | |||||||||||||||||||||||||||||||
| 31/P | O15:K52:H1 | Human | UTI | Spain | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A, K, S, Su, T, Tp |
| 29/P | O15:K52:H1 | Human | UTI | Spain | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A, K, S, Su, T, Tp |
| 28/P | O15:K52:H1 | Human | UTI | Spain | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A, K, S, Su, T, Tp |
| 377 | O15 | Human | UTI | Minneapolis | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | |
| 2H17 | O15:K52:H1 | Human | UTI-blood | Seattle | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | |
| 86.1404 | O15:NM | Turkey | NA | IAb | − | − | + | + | + | II | − | − | − | − | − | − | + | + | − | − | + | + | + | − | − | − | − | − | − | − | K, S, Su, T |
| 86.0041 | O15:NM | Human | NA | NY | F16 | + | + | + | + | II | − | − | − | − | − | + | + | − | − | − | + | − | + | − | − | − | − | − | − | − | |
| AFR017 | O15:HU | Human | UTI-blood | Nairobi | F7-2 | + | + | + | + | II | − | − | − | − | − | + | − | −b | − | − | + | − | + | + | − | − | − | + | − | − | A, C, S, Su, T, Tp |
| CA017 | O15:HU | Human | UTI-blood | Long Beach | F7-2 | + | + | + | + | II | − | − | − | − | − | + | − | −b | − | − | + | − | + | + | − | − | − | + | − | − | A, S, Su, T |
| CA059 | O15:H17 | Human | UTI-blood | Long Beach | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A, C, S, Su, T, Tp |
| 2P9 | O15:K52:H1 | Human | UTI-blood | Seattle | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | S |
| 30/P | O15:K52:H1 | Human | UTI | Spain | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A, K, S, Su, T, Tp |
| CL11A | O15,40,143:H+ | Human | Cystitis | Cleveland | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | |
| 27/P | O15:K52:H1 | Human | UTI | Spain | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | A |
| 1052 | O15 | Human | Pyelo. | Galveston | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | + | − | − | A, C, K, S, Su, T, Tp |
| 8767 | O15:HU | Human | Pyelo. | Galveston | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | T |
| 2607 | O15:HU | Human | Pyelo. | Galveston | F7-2 | + | + | + | + | II | − | − | − | − | − | + | − | −b | − | − | + | − | + | + | − | − | − | + | − | − | A, K, S, Su, T, Tp |
| 6510 | O15:NM | Human | Pyelo. | Galveston | F16 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − | T |
| 4582 | O15:NM | Human | Pyelo. | Galveston | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | A |
| 546 | O15 | Human | UTI | Minneapolis | − | − | − | − | − | − | − | − | + | − | − | + | + | − | − | − | + | − | − | − | − | − | − | − | − | − | A, S, Su, Tp |
| 90.2148 | O15:NM | Human | NA | PA | F16 | + | + | + | + | II | − | − | − | − | − | + | + | − | − | − | + | − | + | − | − | − | − | − | − | − | K, S, Su, T, Tp |
| Non-O15:K52:H1 | |||||||||||||||||||||||||||||||
| 87.0536 | O15:H1 | Human | NA | TX | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | + | − | − | − | − | + | − | − | |
| V32 | O15,40:K+:H1 | Human | UTI-blood | Seattle | F14 | + | + | + | + | III | − | − | − | − | − | − | + | + | − | − | + | − | + | + | + | − | − | + | − | − | A |
| 87.1992 | O15:H1 | Turkey | NA | CA | − | − | + | + | + | II | − | − | − | − | − | − | + | + | − | − | − | + | + | + | − | − | − | + | + | − | A, Su, T |
| 93.0937 | O15 | Water | NA | NY | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | − | − | − | − | + | − | − | A, C, K, S, Su, T, Tp |
| 88.0083 | O15:H1w | Human | NA | DC | F14 | + | + | + | + | III | + | − | − | − | − | − | + | + | − | − | + | − | + | + | + | − | − | + | − | − | A |
| 88.0804 | O15 | Human | NA | MA | F11 | + | + | + | + | II | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | + | K, Su, T |
| 1702 | O15 | Human | Pyelo. | Galveston | F7-1 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | + | + | − | + | − | − | − | − | + | − | + | A |
| 7048 | O15 | Human | Pyelo. | Galveston | F16 | + | + | + | + | II | − | − | + | − | − | + | + | + | − | − | + | − | + | + | − | − | − | + | − | + | A, C, S, Su, T |
| 2659 | O15 | Human | Pyelo. | Galveston | F16 | + | + | + | + | II | − | − | + | − | − | + | + | + | − | − | + | − | + | + | − | − | − | + | − | + | A, C, S, Su, T |
| 95.4449 | O15 | Goose | NA | NY | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | + | − | + | |
| 95.0558 | O15 | Water | NA | NY | F13 | − | + | − | − | − | + | + | − | − | − | − | + | + | − | − | + | + | − | + | − | − | − | − | − | + | |
| 90.0032 | O15:NM | Human | NA | Chile | F10 | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | + | − | + | − | − | − | − | + | − | − | A |
| 97.0041 | O15 | Water | NA | FL | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | + | |
| 87.1732 | O15:NM | Pig | NA | NE | − | − | + | + | + | II | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | K, S, Su, T |
| CL12A | O15,40w:H− | Human | Cystitis | Cleveland | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | + | − | − | + | A |
| 5320 | O15 | Human | Pyelo. | Galveston | F10 | + | + | + | + | II | − | − | − | − | − | + | + | + | − | + | + | − | + | − | − | − | − | + | − | − | A, S |
| 96.0648 | O15:NM | Chicken | NA | NC | − | − | + | + | + | II | − | − | − | − | − | − | + | − | − | − | − | + | + | − | − | − | − | + | + | − | A, S, Su, T, Tp |
| 96.0054 | O15 | Water | NA | NY | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | + | − | − | A, S, Su, T, Tp |
| 98.0335 | O15 | Turkey | NA | NC | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | + | − | − | − | − | + | + | − | |
| 97.0806 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | + | − | − | K, S, Su, T |
| 97.1585 | O15:NM | Cow | NA | CA | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 97.2055 | O15:NM | Pig | NA | NC | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | + | − | − | A, K, S, Su, T |
| 96.0737 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | − | + | − | − | |
| 95.0922 | O15:NM | Chicken | NA | CA | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + | − | − | − | − | + | − | − | C, S, Su, T |
| 95.4297 | O15:Mult | Chicken | NA | Canada (SK) | − | − | + | + | − | II | − | − | − | − | − | − | + | − | − | − | + | + | + | − | − | − | − | + | + | − | Su, T |
| 95.0764 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | + | − | − | T |
| 96.0970 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | T |
| 96.1000 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | T |
| 96.1044 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | T |
| 94.0048 | O15:NM | Human | NA | Zambia | F8 | − | − | − | − | − | − | − | + | − | − | + | + | − | − | − | + | − | + | − | − | − | − | + | − | − | C, Tp |
| 90.2611 | O15:NM | Chicken | NA | PA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | − | + | − | − | − | − | + | − | A, S, Su, T |
| B0S012 | O15:HU | Human | UTI | Boston | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | + | − | − | − | − | − | − | − | − | − | − | |
| 87.0953 | O15 | Chicken | NA | DE | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | + | + | − | − | − | − | + | + | − | |
| 98.1046 | O15 | Turkey | NA | NC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | + | + | − | K, S, T |
| 84.0335 | O15:NM | Pig | NA | NM | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | A, T |
| 90.0318 | O15:NM | Cow | NA | Canada (ON) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | A, S, T |
| 94.0837 | O15 | Water | NA | NY | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | + | − | − | − | + | − | − | S, Su, T |
| 95.4123 | O15:NM | Mink | NA | WA | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | + | − | + | − | − | − | − | + | − | − | A, K, S, Su, T |
| 798 | O15 | Human | UTI | Minneapolis | F10 | − | − | + | − | − | − | − | + | − | − | + | + | −b | − | − | + | − | + | − | − | − | − | + | − | − | A, K, S, Su, T, Tp |
| 87.0276 | O15 | Cow | NA | IA | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | − | − | − | + | − | − | + | − | + | − | − | |
| 86.0512 | O15:NM | Rabbit | NA | MO | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | + | − | + | − | − | − | − | + | − | − | |
| 91.1135 | O15:NM | Human | NA | Saudi Arabia | F10 | − | − | + | − | − | − | − | + | − | − | + | + | − | − | − | + | − | + | − | − | − | − | + | − | − | A, K, S, Su, T, Tp |
| 88.0279 | O15:NM | Horse | NA | AL | − | − | − | − | − | − | + | + | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − | − | − | − | A, C, K, S, Su, T, Tp |
| 91.0796 | O15:NM | Dog | NA | WY | F12 | + | + | + | − | − | + | + | − | − | − | − | + | −b | − | − | + | − | + | + | − | − | − | + | − | − | |
| 96.0953 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | T |
| 96.1723 | O15 | Cow | NA | MI | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 97.1871 | O15 | Chicken | NA | ND | − | − | + | + | + | II | − | − | − | − | − | − | + | − | − | − | + | − | + | − | − | − | − | + | − | − | K, S, Su, T |
| 87.0637 | O15:NM | Dog | NA | PA | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | Su |
| 93.1226 | O15:NM | Deer | NA | CA | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | |
| 88.1341 | O15:NM | Horse | NA | KS | − | − | − | − | − | − | + | + | − | + | + | − | + | + | − | − | + | + | + | − | − | − | − | − | − | − | |
| 88.0998 | O15 | Rabbit | NA | Hungary | − | − | − | − | − | − | + | − | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | + | − | − | |
| 85.0706 | O15 | Chicken | NA | NH | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | − | − | − | − | − | − | − | T |
| 98.0874 | O15 | Turkey | NA | NC | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + | − | − | − | − | + | + | − | Su |
| CL38 | O15 | Human | Cystitis | Cleveland | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | A |
| CL31 | O15,40,143:NM | Human | Cystitis | Cleveland | F10 | − | − | + | − | − | − | − | + | − | − | + | + | −b | − | − | + | − | + | − | − | − | − | + | − | − | A, S, Su, T, Tp |
| CL29 | O15,40,143:NM | Human | Cystitis | Cleveland | F10 | − | − | + | − | − | − | − | + | − | − | + | + | −b | − | − | + | − | + | − | − | − | − | + | − | − | A, T |
| 6081 | O15 | Human | Pyelo. | Galveston | F10 | − | − | + | − | − | − | − | + | − | − | − | + | −b | − | − | + | − | − | − | − | − | − | + | − | − | T |
| 86.0627 | O15:NM | Chicken | NA | DE | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | S |
| BOS007 | O15 | Human | Blood | Boston | F14 | + | + | + | + | III | − | − | − | − | − | − | + | − | − | − | + | − | + | + | + | − | − | + | − | − | |
Strains are listed in the same sequence as in the PCR-based dendrogram (Fig. 1), with members of the O15:K52:H1 clonal group shown first. Strains were variously typed for the complete O: K: H serotype, the O: H serotype, or the O antigen only; all initially available seroantigens are shown. Plus signs indicate presence of a trait, minus signs absence of a trait. Abbreviations: Pyelo, pyelonephritis; UTI-blood, urosepsis; Abd./blood, abdominal source bacteremia; NA, not available or not applicable; sfa/foc, S and FIC fimbriae, focG, putative FIC adhesin; afa/dra, afimbrial adhesins and Dr-binding adhesins; bmaE, M fimbriae; gafD, G fimbriae; iha, iron-regulated gene homologue adhesin; fimH, type 1 fimbrial adhesin molecule; kpsMT, capsule synthesis (group II, group III, or K1 variant within group II; “−b” indicates detection of kpsMT II by blot but not PCR, consistent with presence of the K2 kpsMT variant); fyuA, yersiniabactin receptor; iroN, putative siderophore receptor; iutA, aerobactin receptor; hlyA, hemolysin; cnf1, cytotoxic necrotizing factor 1; cdtB, cytolethal distending toxin; ibeA, invasion of brain endothelium protein Ibe10; traT, serum resistance-associated gene; cvaC, colicin V; malX, pathogenicity associated island marker from pyelonephritis isolate CFT073. Strains shown as positive for a specific papA allele (i.e., F type) but not for papA failed to amplify with the (flanking) papAH primers but were positive in the F PCR assay for an allele-specific subregion within papA. All strains were negative by PCR for sfaS (S fimbrial adhesin) and nfaE (nonfimbrial adhesin). Antibiotics: A, ampicillin; C, chloramphenicol; K, kanamycin; S, streptomycin; Su, sulfamethoxazole; T, tetracycline; Tp, trimethoprim.
Two-letter abbreviations are used for the states within the United States.
fliC variant analysis.
Detection of the flagellin gene fliC and discrimination between its H antigen-specific variants was done for all non-European members of the O15:K52:H1 clonal group by PCR-restriction fragment length polymorphism (RFLP) analysis, as described by Fields et al. (7). Consensus primers specific for conserved regions at the extreme 5′ and 3′ bounds of fliC were used to amplify the entirety of fliC. The resulting fliC amplicons were then restricted with RsaI to generate variant-specific RFLP patterns. One of the Spanish O15:K52:H1 controls was used for a reference.
Antimicrobial resistance profiles.
Susceptibility to seven antimicrobial agents was determined by a disk diffusion method with Mueller-Hinton agar and commercial antibiotic-impregnated disks (Difco, Detroit, Mich.) (30). The agents assayed were ampicillin (A), chloramphenicol (C), kanamycin (K), streptomycin (S), sulfamethoxazole (Su), tetracycline (T), and trimethoprim (Tp). Procedures and interpretive criteria were as proposed for Enterobacteriaceae by the National Committee for Clinical Laboratory Standards (31). The antibiotic resistance score was the number of agents to which a strain was found to be resistant or intermediately susceptible (16).
Conjugation experiments.
Conjugation experiments were done to determine whether in certain strains antimicrobial resistance determinants were carried on transferable plasmids containing aerobactin-encoding genes, as was previously reported for the E. coli O15:K52:H1 isolates from the South London outbreak of 1986 to 1987 (36; S. J. Eykyn and I. Phillips, Letter, Lancet ii:1454, 1986; Wright and Perinpanayagam, letter). Among the 16 non-European members of the O15:K52:H1 clonal group from the present study, the 10 isolates that were positive for iutA and exhibited resistance to at least one of the antibiotics studied were screened for the ability to coordinately transfer antibiotic resistance and iutA to an E. coli K-12 recipient (16). Positive controls included two (iutA positive, multiply antibiotic resistant) European representatives of the O15:K52:H1 clonal group (one from Barcelona, Spain; one from London, United Kingdom) (38) and two non-O15:K52:H1 urosepsis isolates (H27 and V10) from Seattle, Wash., that contain aerobactin-encoding plasmids and exhibit multiple antibiotic resistance (16).
Donor strains were mated with nalidixic acid-resistant K-12 recipient strains 395-1 (provided by Steve Moseley, University of Washington, Seattle) (40) or JM109 (Promega, Madison, Wis.), which have no plasmids and are PCR negative for iutA (not shown), by using a broth mating procedure, as previously described (8, 16). Putative transconjugants were selected on Luria agar supplemented with nalidixic acid at 50 μg per ml, plus (separately) each of the antimicrobial agents to which the donor strain was resistant, in concentrations appropriate for the particular agents (28). From the mating mixtures, putative transconjugants that exhibited resistance to both nalidixic acid and the second agent were compared with both parents by RAPD analysis (primer 1290) (1) to confirm the genomic identity with the recipient strain. Confirmed transconjugants were then tested by PCR for acquisition of iutA (23).
Statistical methods.
Comparison of proportions were tested by using Fisher's exact test. Comparisons of prevalence within the same population were tested by using McNemar's test (7). Comparisons of antibiotic resistance scores were tested by using a two-tailed t test. Because of the multiple comparisons, P ≤ 0.01 was used as the threshold for statistical significance, with P < 0.05 considered to reflect borderline statistical significance. Similarity relationships with respect to composite VF profiles (independent of phylogenetic background) were assessed by constructing a dendrogram according to UPGMA by using the application NTSYS (Exeter Software, Stony Brook, N.Y.).
RESULTS
Phylogenetic analysis and confirmatory serotyping.
Cluster analysis of composite RAPD and rep-PCR profiles for the five Spanish and 75 non-European O15 isolates (80 isolates total) resolved four major clusters, or putative phylogenetic groups. Within one of these clusters, the predominant subcluster contained all five of the Spanish O15:K52:H1 controls and hence was considered to represent the O15:K52:H1 clonal group (Fig. 1). Accordingly, the 16 non-European strains in this subcluster, which included the two urosepsis isolates from Seattle that were already known to be of serotype O15:K52:H1, were defined as members of the O15:K52:H1 clonal group (Fig. 1).
Epidemiological and geographic associations.
Among the 75 non-European O15 isolates, membership in the O15:K52:H1 clonal group was significantly associated with isolation from a human host. Fifteen (94%) of the 16 clonal group members, but only 19 (32%) of the 59 nonclonal group members, were known to have been isolated from humans (P < 0.001). The sole nonhuman isolate in this group was from a turkey. The 15 human isolates represented diverse host populations, including pregnant and nonpregnant women, men, and children and were from various extraintestinal infection syndromes, including pyelonephritis, cystitis, urosepsis, and abdominal-source sepsis (Table 1). They accounted for 44% of the 34 known human isolates in the non-European study population and for 13 (50%) of the 26 serogroup O15 isolates from the five defined non-European human clinical collections, i.e., for 2.8% of all E. coli isolates from those collections. The non-European clonal group members derived from diverse geographical locales, including cities throughout the United States (Cleveland, Galveston, Long Beach, Minneapolis, New York, and Seattle) and Nairobi, Kenya (Table 1).
VF genotypes (Tables 1 and 2).
TABLE 2.
Distribution of virulence factors among members of the O15:K52:H1 clonal group versus other O15 strains
| Associated traita | Prevalence of associated trait amongb:
|
Pc
|
|||
|---|---|---|---|---|---|
| Spanish O15:K52:H1 isolates (n = 5) | Non-European O15:K52:H1 clonal group isolates (n = 16) | Other O15 members (n = 59) | A vs C | B vs C | |
| papA | 5 (100) | 13 (81) | 9 (15) | <0.001 | <0.001 |
| F16 | 5 (100) | 10 (63) | 2 (3) | <0.001 | <0.001 |
| F7-2 | 0 | 3 (19) | 0 | 0.007 | |
| F10 | 0 | 0 | 7 (12) | ||
| papC | 5 (100) | 14 (88) | 15 (25) | 0.002 | <0.001 |
| papEF | 5 (100) | 14 (88) | 20 (34) | 0.007 | <0.001 |
| papG | 5 (100) | 14 (88) | 12 (20) | 0.001 | <0.001 |
| Allele II | 5 (100) | 14 (88) | 10 (17) | <0.001 | <0.001 |
| Allele III | 0 | 0 | 3 (5) | ||
| sfa/focDE | 0 | 0 | 6 (10) | ||
| focG | 0 | 0 | 4 (7) | ||
| afa/draBC | 0 | 1 (6) | 9 (15) | ||
| bmaE | 0 | 0 | 1 (2) | ||
| gafD | 0 | 0 | 3 (5) | ||
| iha | 5 (100) | 14 (88) | 13 (22) | 0.001 | <0.001 |
| fimH | 5 (100) | 13 (81) | 53 (90) | ||
| kpsMT II | |||||
| PCR | 5 (100) | 9 (56) | 18 (30) | 0.004 | |
| Blot | 5 (100) | 12 (75) | 23 (39) | 0.01 | 0.01 |
| K1 | 0 | 0 | 3 (5) | ||
| K2 | 0 | 3 (18) | 5 (9) | ||
| kpsMT III | 0 | 0 | 5 (8) | ||
| fyuA | 5 (100) | 16 (100) | 33 (56) | <0.001 | |
| iroN | 0 | 1 (7) | 10 (17) | ||
| iutA | 5 (100) | 14 (88) | 29 (49) | 0.01 | |
| hlyA | 0 | 3 (19) | 10 (17) | ||
| cnf1 | 0 | 0 | 3 (5) | ||
| cdtB | 0 | 0 | 1 (2) | ||
| ibeA | 0 | 0 | 1 (2) | ||
| traT | 0 | 4 (25) | 43 (73) | 0.003 | 0.002 |
| cvaC | 0 | 0 | 8 (13) | ||
| malX | 0 | 0 | 8 (13) | ||
pap, P fimbriae; F16, F7-2, and F10, alleles of papA (structural subunit); alleles II and III, papG (adhesin) variants; sfa/foc, S and F1C fimbriae; focG, F1C fimbriae; afa/dra, Dr-binding adhesins; bma, M fimbriae; gaf, G fimbriae; iha, iron-regulated gene-homologue adhesin; fimH, type 1 fimbrial adhesin; kpsMT II, group II capsule (as detected by PCR or blot), including the K1 and K2 variants; kpsMT III, group III capsule; fyuA, yersiniabactin receptor; iroN, putative siderophore receptor, iutA, aerobactin receptor, hly, hemolysin; cnf1, cytotoxic necrotizing factor 1; cdtB, cytolethal distending toxin; ibeA, invasion of brain endothelium A; traT, serum resistance-associated; cvaC, colicin V; malX, marker for pathogenicity island from strain CFT073.
Values are numbers of isolates. Values in parentheses are percentages.
A, B, and C are the Spanish O15:K52:H1 control isolates, non-European O15:K52:H1 clonal group members, and nonclonal group O15 isolates, respectively. P values shown where <0.05. For all comparisons of A versus B, P > 0.10.
Certain VF genes were significantly more prevalent among the five Spanish O15:K52:H1 controls and/or the 16 non-European O15:K52:H1 clonal group members than among the 59 nonclonal group O15 isolates. The O15:K52:H1-associated VF genes included all four pap regions, the F7-2 and F16 papA alleles, papG allele II, iha (iron-regulated gene homologue adhesin), kpsMT II (group 2 capsules [by blot and PCR]), fyuA (yersiniabactin receptor), and iutA (aerobactin receptor). In contrast, traT (associated with serum resistance) was negatively associated with the O15:K52:H1 clonal group (Tables 1 and 2). The Spanish and the non-European O15:K52:H1 clonal group members did not differ significantly with respect to the prevalence of any of the studied VFs (Table 2).
Only three of the O15:K52:H1 clonal group members contained a papA allele other than the F16 allele. These three strains (AFR017, CA017, and 2607; from Nairobi, Long Beach, and Galveston, respectively) all exhibited the F7-2 papA allele. They also differed from other O15:K52:H1 clonal group members by exhibiting the K2 variant of kpsMT II and containing hlyA and traT (Table 1). Conversely, only two non-O15:K52:H1 clonal group members contained the F16 papA allele. Both of these strains (strains 7048 and 2659, both from Galveston) differed from the F16-positive O15:K52:H1 clonal group members by containing afa/dra (Dr-binding adhesins), hlyA, traT, and malX (Table 1).
Like three of the O15:K52:H1 clonal group members, 5 of the 59 non-O15:K52:H1 clonal group members (strains 798, 91.0796, CL31, CL29, and 6081) exhibited the K2 variant of kpsMT II (Table 1). However, these strains exhibited VF profiles that were otherwise quite dissimilar from those of the K2 variant-positive clonal group members. Four of the K2 variant-positive non-O15:K52:H1 clonal group isolates exhibited the F10 (rather than the F16) papA allele, were otherwise pap-negative except for papEF, and contained afa/dra, whereas the remaining strain (isolate 91.0796) exhibited the F12 papA allele, sfa/foc, focG, and hly (Table 1).
Cluster analysis of VF profiles.
Correspondence of VF profiles with phylogenetic background was further assessed by using UPGMA to generate a dendrogram based on the VF data from Table 1. This dendrogram showed the population to segregate into two well-separated clusters: one of which contained 33 largely VF-positive isolates, the other 47 were largely VF-negative isolates (not shown). Within the first (VF-positive) cluster, the predominant subcluster comprised 20 isolates, 17 of which (5 Spanish, 13 non-European) were members of the O15:K52:H1 clonal group (P < 0.001 for the association of this subcluster with the O15:K52:H1 clonal group). This analysis confirmed statistically both the high degree of homogeneity among the O15:K52:H1 clonal group members and the nearly complete segregation of these strains from other O15 isolates with respect to VF profiles.
fliC PCR-RFLP analysis.
All 16 non-European O15:K52:H1 clonal group members and 1 of the Spanish O15:K52:H1 controls (strain 31/P) underwent RsaI PCR-RFLP analysis of the flagellin gene fliC. Thirteen of the sixteen non-European isolates (including all seven that were initially serotyped as H1, two that initially had not been H serotyped, two that were initially reported as nonmotile, and one that was initially serotyped as H17: Table 1), plus the Spanish O15:K52:H1 control, exhibited identical fliC RFLP profiles, designated “pattern A” (as shown for selected isolates in Table 3). This consensus profile corresponded with the H1 fliC profile as described by Fields et al. (6). The three remaining non-European clonal group members (AFR017 and CA017, both initially reported as H untypeable, and 2607, initially not H serotyped) all exhibited a variant VF profile that included the F7-2 papA allele, the K2 kpsMT variant, and hly (Table 1). All three isolates exhibited an identical fliC PCR-RFLP pattern (“pattern B”), one that was distinct from the H1 pattern (as shown for CA017 and 2607 in Table 3).
TABLE 3.
Seroantigens and other characteristics of selected non-European representatives of the O15:K52:H1 clonal group
| Strain | O antigen or serotypea
|
fliCb | Hostc | Syndromed | Location | Virulence genotypee
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial | Confirmatory | F type | papA | papC | papG | afa | iha | fimH | kpsMT | K2 | iroN | iutA | hly | traT | |||||
| 377 | O15 | O15:K52:H1 | A | Woman | UTI | Minneapolis | F16 | + | + | II | − | + | + | II (P,b) | − | − | + | − | − |
| CL11A | O15,40,143:H+ | O15:K52:H18 | A | Child | Cystitis | Cleveland | F16 | + | + | II | − | + | + | II (P,b) | − | − | + | − | − |
| 1052 | O15 | O15:K52:H1 | A | Pregnant | G pyelo. | Galveston | F16 | + | + | II | − | + | + | II (P,b) | − | − | + | − | + |
| 90.2148 | O15:NM | O15:K52:H1 | A | Human | NA | Pennsylvania | F16 | + | + | II | − | + | + | II (P,b) | − | − | + | − | − |
| CA017 | O15:HU | O15:K2:H18 | B | Adult | Urosepsis | Long Beach | F7-2 | + | + | II | − | + | − | II (b) | + | − | + | + | + |
| 2607 | O15:HU | O15:K2:H18 | B | Pregnant | G pyelo. | Galveston | F7-2 | + | + | II | − | + | − | II (b) | + | − | + | + | + |
| 4582 | O15:NM | O15:K16:H1 | A | Pregnant | G pyelo. | Galveston | − | − | − | − | − | − | + | II (P,b) | − | − | − | − | − |
| 546 | O15 | O15:K16:H1 | A | Woman | UTI | Minneapolis | − | − | − | − | + | + | + | II (P,b) | − | − | − | − | − |
| 86.1404 | O15:NM | O15:K52:H− | A | Turkey | NA | Iowa | − | − | + | II | − | − | + | II (P,b) | − | + | + | − | − |
Initial, as available prior to study. Confirmatory, as newly determined for present study. H+ and HU, motile but H untypeable; H− and NM, nonmotile; K+, K untypeable.
fliC PCR-RFLP pattern.
Pregnant hosts were women; adult host was human.
UTI, urinary tract infection; G pyelo., gestational pyelonephritis; NA, not available.
Only those virulence markers for which the nine strains exhibited nonidentity are shown. F type refers to papA allele. Results for papEF were as for papC. papG refers to papG allele. For kpsMT, P and b refer to PCR and blot, respectively. K2 status inferred from combined PCR and blot result for kpsMT II.
Confirmatory O:K:H serotyping.
Complete O:K:H serotypes were determined by the WHO Laboratory in Copenhagen for nine of the newly defined non-European members of the O15:K52:H1 clonal group for which only O:H serotypes or O antigens were previously known. These isolates were selected so as to include representatives of all unique O:H serotypes and VF profiles encountered among the putative clonal group members (Table 3). All nine isolates were confirmed as O15 positive. Various K and H antigens were identified, in patterns corresponding with the VF profile in a manner suggesting the existence of discrete subclones within the larger clonal group (Table 3). Five isolates, four of which were F16 papA positive and one of which lacked papA, were positive for the K52 capsular antigen. Three of these tested as H1, one as H18, and one was nonmotile. In contrast, both of the F7-2 papA-positive isolates that underwent confirmatory serotyping were reported as K2:H18, and both pap-negative isolates were reported as K16:H1 (Table 3). Precise correspondence was observed between confirmatory H antigen results and fliC PCR-RFLP patterns for all strains that underwent confirmatory serotyping excepting CL11A, which was serotyped as H18 but which yielded the H1-associated fliC RFLP pattern (pattern “A” in Table 3).
Antimicrobial resistance.
Resistance to one or more of the seven antimicrobial agents tested (A, C, K, S, Su, T, and Tp) was variably present within the population (Table 1). In contrast to the VF profiles, antimicrobial resistance patterns corresponded minimally with phylogenetic group, host group, or geographical origin. The distinctive “ACSSuTTp” antimicrobial resistance profile of the South London epidemic strain occurred in only two (13%) of the non-European O15:K52:H1 clonal group members (strains AFR017, from Nairobi, and CA059, from Long Beach) but in none of the 59 nonclonal group O15 isolates (P = 0.04). Antimicrobial resistance scores were significantly higher among the Spanish O15:K52:H1 controls (mean, 4.2) than among the 59 nonclonal group O15 isolates (mean, 2.1; P = 0.028) but not than among the 16 non-European O15:K52:H1 clonal group members (mean, 2.9; P > 0.10). Resistance scores did not differ significantly between the two non-European populations (P > 0.10).
Conjugation experiments.
The multiple antimicrobial resistance phenotype of many of the O15:K52:H1 strains, together with the high prevalence of iutA, suggested that within the O15:K52:H1 clonal group antimicrobial resistance determinants might be carried on transferable plasmids containing aerobactin-encoding genes, as was previously documented for the South London epidemic strain (36). To test this hypothesis, attempts were made to coordinately transfer iutA and antimicrobial resistance markers by conjugation from selected iutA-positive non-European O15:K52:H1 clonal group donors to an E. coli K-12 recipient. Although it was possible to transfer resistance to tetracycline, streptomycin, ampicillin, and/or kanamycin from 6 of the 10 non-European O15:K52:H1 donors tested (strains 6510, 1052, 8767, 90-2148, and 86-1404), none of the resulting transconjugants were positive for iutA (not shown). In contrast, among the mating controls, coordinate transfer of antibiotic resistance and iutA was achieved with both of the European O15:K52:H1 strains and with both American non-O15:K52:H1 urosepsis isolates (not shown).
DISCUSSION
In the present study we compared 75 non-European E. coli isolates of serogroup O15 with five representatives of the O15:K52:H1 clonal group from Barcelona, Spain, with respect to genomic background, extended virulence genotypes, and antimicrobial resistance profiles. We found that, based on phylogenetic criteria, 16 (21%) of the non-European O15 isolates belonged to the O15:K52:H1 clonal group, which heretofore has been described only in Europe. Non-European clonal group members represented diverse geographical origins, were isolated almost exclusively from humans, and accounted for 50% of the O15 isolates from five defined human clinical collections. They exhibited a narrow range of distinctive VF profiles which corresponded closely with those of the Spanish O15:K52:H1 controls but displayed diverse antimicrobial resistance patterns that were largely indistinguishable from those of the nonclonal group O15 isolates. Certain members of the clonal group expressed alternative surface antigens (K16, K2, and/or H18) not previously associated with this clonal group.
Our findings provide the first genetic evidence of the presence of the O15:K52:H1 clonal group outside of Europe (32, 33, 36, 38; Dalmau et al., letter; Eykyn and Phillips, letter; Reeves et al., letter; Waghorn et al., letter; Wright and Perinpanayagam, letter). The validity of the PCR-based phylotyping is supported by the placement within the same cluster of all five Spanish O15:K52:H1 control strains, which were previously shown to be clonally related according to ribotyping and biotyping (38), and by the placement within this cluster also of the two urosepsis isolates from Seattle which were already known to be of serotype O15:K52:H1 (21, 23). Further support is provided by detection of the K52 capsular antigen and the H1 flagellar antigen (n = 5 each) among the nine newly identified putative clonal group members that underwent confirmatory O:K:H serotyping.
The O15:K52:H1 clonal group was found to account for half of the serogroup O15 isolates from the five defined clinical collections of human extraintestinal infection isolates that contributed to the present study population and for 2.8% of all isolates from these collections. The latter value is similar to the reported prevalence of this clonal group among urine and blood isolates from Barcelona, Spain (1.3 and 2.9%, respectively) (38), and among blood isolates from Copenhagen, Denmark (4.5%) (33) and Terrassa, Spain (4.4%) (Dalmau et al., letter). Taken together, these observations suggest that the O15:K52:H1 clonal group is globally distributed and makes an appreciable, albeit minor (as with most individual clonal groups), contribution to endemic extraintestinal infections in humans, quite apart from its unusual epidemic behavior during the South London outbreak of 1986 to 1987 (32, 36; Eykyn and Phillips, letter; Waghorn et al., letter; Wright and Perinpanayagam, letter). It thus qualifies as an extraintestinal pathogenic E. coli (ExPEC) clone (38, 39).
The pathogenic versatility of the O15:K52:H1 clonal group is evidenced by the clone's participation in diverse extraintestinal infection syndromes in varied host populations (Table 1). The apparent anthropotropism of the clonal group, which is suggested by the group's strong association with human as opposed to animal or environmental sources, has at least two possible explanations. First, clonal group members may be better adapted for causing extraintestinal infections in humans than are other O15 strains. This would be consistent with their possession of specific virulence traits known to be associated with human extraintestinal infections (Table 2) (10, 22, 23, 39). Alternatively, clonal group members may be more prevalent in the human environment, including possibly the human gut flora, than are other O15 strains, and hence may have more opportunity for causing infections in humans than in animals, whereas the reverse may be true for other O15 strains. Further study would be needed to select between these “special pathogenicity” and “prevalence” hypotheses.
The diversity of antimicrobial resistance patterns noted within the O15:K52:H1 clonal group, and the general absence of resistance profile differences between these strains and the nonclonal group O15 isolates, is consistent with previous observations involving European representatives of the O15:K52:H1 clonal group, which exhibited greater diversity with respect to antimicrobial resistance than for any other measured trait (38). Diversity of resistance patterns is likely attributable to the varied selection environments and opportunities for acquisition of resistance elements to which the various O15 strains have been exposed, combined with the ease with which resistance determinants (as opposed to virulence genes and other genomic elements) can be exchanged between strains. Thus, although the “ACSSuTTp” resistance profile was a useful marker for the outbreak strain during the 1986 to 1987 South London epidemic (32, 36; Eykyn and Phillips, letter; Waghorn et al., letter; Wright and Perinpanayagam, letter), it clearly cannot be relied on to identify the O15:K52:H1 clonal group in other contexts.
Indeed, among recent acute cystitis and pyelonephritis isolates in the United States, the ACSSuTTp resistance pattern is highly predictive of “clonal group A” (20, 27). Clonal group A is closely related phylogenetically to the O15:K52:H1 clonal group and exhibits essentially the same VF profile but is considerably less diverse with respect to resistance profiles (20, 27, 38). The comparative homogeneity of resistance profiles in clonal group A may be attributable to the more recent emergence and expansion of clonal group A, compared with the O15:K52:H1 clonal group (20, 27). Of note, our finding of the H18 flagellar antigen in certain non-European members of the O15:K52:H1 clonal group provides another point of commonality between this clonal group and clonal group A, the members of which characteristically exhibit serotypes O11:K52:H18, O17:K52:H18, or O77:K52:H18 (20). Further study is warranted regarding the evolutionary relationships between these two recently recognized epidemic-associated clonal groups from phylogenetic group D.
The demonstrated conjugative transfer of resistance markers from certain clonal group members into E. coli K-12 suggests that antimicrobial resistant members of the O15:K52:H1 clonal group may provide a source of resistance elements for acquisition by other strains. In certain European members of this clonal group (and in certain other strains) antimicrobial resistance elements can be transferred horizontally on plasmids containing aerobactin-encoding genes (16, 36). However, in the present study attempts to demonstrate coordinate transfer of the aerobactin system (iutA) and resistance markers among non-European clonal group members were unsuccessful. Moreover, previous genetic analysis of the two O15:K52:H1 urosepsis isolates from Seattle indicated that these strains have a chromosomal rather than a plasmid aerobactin system (16). These findings suggest that in different subsets of the O15:K52:H1 clonal group not only may varied antimicrobial resistance elements be present (Table 1) but similar resistance markers may occur in different genetic environments and/or have different modes of horizontal transfer. In this regard, the four Spanish clonal group members included in the present study that are multiply resistant (28/P, 29/P, 30/P, and 31/P) also are positive for class I integrons by PCR (26), whereas susceptible strain 27/P is negative (G. Prats, unpublished data), evidence consistent with possible integron-mediated acquisition and loss of resistance markers in some members of the clonal group.
Group D is the major E. coli phylogenetic group other than group B2 that exhibits a high concentration of extraintestinal VFs (3, 13, 37). It also is the origin of certain familiar virulent ExPEC clones such as O1:K1:H−, O7:K1:H−, O157:K+, and O2:K5:H4 (13, 18). The previous placement of the O15:K52:H1 clonal group within group D, as inferred based on comparative RAPD analysis of clonal group members and selected ECOR strains (20, 38), was indirectly confirmed in the present study. Non-European bacteremia isolates AFR017, CA017, and CA059, which were shown in the present study to be members of the O15:K52:H1 clonal group, derive from cluster V of Maslow et al. (29), which has been shown through comparative RAPD analysis to correspond with ECOR phylogenetic group D (18). In addition, we confirmed the phylogenetic group D origin of this clonal group by a group-specific multiplex PCR assay (4) (unpublished data). It should be noted that the VF profiles of most O15:K52:H1 clonal group members are typical of E. coli phylogenetic group D, including positivity for pap, papG allele II, and kpsMT II but the absence of papG allele III, hly, sfa, and cnf (2, 3, 13, 18, 37).
In summary, we found that the O15:K52:H1 clonal group, which recently has been shown to merit consideration as a uropathogenic clone in Europe, is broadly disseminated also in the United States, where it preferentially infects humans and accounts for ca. 50% of human extraintestinal E. coli isolates of serogroup O15. The consensus VF profile of non-European members of the O15:K52:H1 clonal group, which is similar to that of European members of this clonal group and to that of closely related clonal group A, includes the F16 papA allele, papG allele II, iha, kpsMT II, fyuA, and iutA. Occasional isolates lack most or all of these traits, whereas others exhibit instead the F7-2 papA allele, hlyA, and traT, and lack kpsMT II. Antibiotic resistance profiles are highly variable. Heightened awareness of this virulent and anthropotropic clonal group should lead to improved understandings of its prevalence, distribution, clinical associations, pathogenetic mechanisms, and evolving patterns of antimicrobial resistance.
Acknowledgments
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs VA Merit Review (J.R.J. and J.N.M.), National Institutes of Health grants DK-47504 (J.R.J) and DK-42029 (B.N.), National Research Initiative Competitive Grants Program/U.S. Department of Agriculture grant 00-35212-9408 (J.R.J.), and grant 95/1379 from the Fondo de Investigaciones Sanitarias de la Seguridad de España (G.P.).
Dave Prentiss helped prepare Fig. 1, and Ann Emery helped prepare the manuscript (both are from the Minneapolis VA Medical Center).
REFERENCES
- 1.Berg, D. E., N. S. Akopyants, and D. Kersulyte. 1994. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol. Cell. Biol. 5:13-24. [Google Scholar]
- 2.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., and D. L. Hartl. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjardins, P., B. Picard, B. Kaltenbock, J. Elion, and E. Denamur. 1995. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 41:440-448. [DOI] [PubMed] [Google Scholar]
- 6.Fields, P. I., K. Blom, H. J. Hughes, L. O. Helsel, P. Feng, and B. Swaminathan. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J. Clin. Microbiol. 35:1066-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleiss, J. L. 1981. Statistical methods for rates and proportions, p. 112-137. John Wiley & Sons, New York, N.Y.
- 8.Hardy, K. 1983. Bacterial plasmids. American Society for Microbiology, Washington, D.C.
- 9.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNS-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, J. R., J. J. Brown, and J. N. Maslow. 1998. Clonal distribution of the three alleles of the Gal(α1-4)Gal-specific adhesin gene papG among Escherichia coli strains from patients with bacteremia. J. Infect. Dis. 177:651-661. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, J. R., and C. Clabots. 2000. Improved repetitive element-(rep-) polymerase chain reaction fingerprinting of Salmonella with the use of extremely elevated annealing temperatures. Clin. Diagn. Lab. Immunol. 7:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J. R., P. Delavari, and T. O'Bryan. 2001. Escherichia coli O18:K1:H7 isolates from acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J. Infect. Dis. 183:425-434. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, J. R., C. E. Johnson, and J. N. Maslow. 1999. Clinical and bacteriologic correlates of the papG alleles among Escherichia coli strains from children with acute cystitis. Pediatr. Infect. Dis. J. 18:446-451. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, J. R., S. Moseley, P. Roberts, and W. E. Stamm. 1988. Aerobactin and other virulence factor genes among strains of Escherichia coli causing urosepsis: association with patient characteristics. Infect. Immun. 56:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. R., and T. T. O'Bryan. 2000. Improved repetitive element (rep-)-polymerase chain reaction (rep-PCR) fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Diagn. Lab. Immunol. 7:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. R., T. T. O'Bryan, M. A. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. R., T. T. O'Bryan, D. A. Low, G. Ling, P. Delavari, C. Fasching, T. A. Russo, U. Carlino, and A. L. Stell. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli that express papG allele III. Infect. Immun. 68:3327-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., T. T. O'Bryan, A. R. Manges, and L. W. Riley. 2001. Emergence of a multi-drug resistant clonal groups of extraintestinal pathogenic Escherichia coli as a significant cause of acute pyelonephritis. Clin. Infect. Dis. 33:1234. [Google Scholar]
- 21.Johnson, J. R., I. Orskov, F. Orskov, P. Goullet, B. Picard, S. L. Moseley, P. L. Roberts, and W. E. Stamm. 1994. O, K, and H antigens predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J. Infect. Dis. 169:119-126. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. J. Vary, and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, J. R., A. L. Stell, F. Scheutz, T. T. O'Bryan, T. A. Russo, U. B. Carlino, C. C. Fasching, J. Kavle, L. van Dijk, and W. Gaastra. 2000. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex polymerase chain reactions-based assay. Infect. Immun. 68:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 26.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Maslow, J. N., T. S. Whittam, C. F. Gilks, R. A. Wilson, M. E. Mulligan, K. S. Adams, and R. D. Arbeit. 1995. Clonal relationships among bloodstream isolates of Escherichia coli. Infect. Immun. 63:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Committee for Clinical and Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 7th ed. (M2-A7), vol. M2-A7, p. 1-18. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.National Committee for Clinical and Laboratory Standards. 2000. Supplemental tables, p. 14-18. Disk diffusion. M100-S10 (M2). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 32.O'Neill, P. M., C. A. Talboys, A. P. Roberts, and B. S. Azadian. 1990. The rise and fall of Escherichia coli O15 in a London teaching hospital. J. Med. Microbiol. 33:23-27. [DOI] [PubMed] [Google Scholar]
- 33.Olesen, B., H. J. Kolmos, F. Orskov, and I. Orskov. 1995. A comparative study of nosocomial and community-acquired strains of Escherichia coli causing bacteraemia in a Danish university hospital. J. Hosp. Infect. 31:295-305. [DOI] [PubMed] [Google Scholar]
- 34.Orskov, I., and F. Orskov. 1985. Escherichia coli in extra-intestinal infections. J. Hyg. 95:551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orskov, I., F. Orskov, A. Birch-Andersen, M. Kanamori, and C. Svanborg Eden. 1982. O, K, H and fimbrial antigens in Escherichia coli serotypes associated with pyelonephritis and cystitis. Scand. J. Infect. Dis. 33:18-25. [PubMed] [Google Scholar]
- 36.Phillips, I., S. Eykyn, A. King, W. R. Grandsden, B. Rowe, J. A. Frost, and R. J. Gross. 1988. Epidemic multiresistant Escherichia coli infection in West Lambeth health district. Lancet i:1038-1041. [DOI] [PubMed]
- 37.Picard, B., J. Sevali Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prats, G., F. Navarro, B. Mirelis, D. Dalmau, N. Margall, P. Coll, A. Stell, and J. R. Johnson. 2000. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J. Clin. Microbiol. 38:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo, T. A., and J. R. Johnson. 2000. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 40.Sansonetti, P. J., T. L. Hale, G. J. Dammin, C. Kapfer, H. H. J. Collins, and S. B. Formal. 1983. Alterations in the pathogenecity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun. 39:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokal, R. R., and P. H. A. Sneath. 1963. Principles of numerical taxonomy, W. H. Freeman Co., San Francisco, Calif.
- 42.Tullus, K., K. Horlin, S. B. Svenson, and G. Källenius. 1984. Epidemic outbreaks of acute pyelonephritis caused by nosocomial spread of P fimbriated Escherichia coli in children. J. Infect. Dis. 150:728-736. [DOI] [PubMed] [Google Scholar]
- 43.Versalovic, J., M. Schneid, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 44.Wang, G., T. S. Whittam, C. M. Berg, and D. E. Berg. 1993. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 21:5930-5933. [DOI] [PMC free article] [PubMed] [Google Scholar]