Abstract
To identify ehrlichial agents in Boophilus microplus ticks, DNA samples of B. microplus collected from the Tibet Autonomous Region and Sichuan Province of China were screened by a nested PCR. Sixteen of 43 (37%) DNA samples of B. microplus from Tibet were positive in nested PCR analysis. All 27 samples from Sichuan were negative. The screen identified two ehrlichial agents based on different 16S rRNA genes that were found after amplifying and sequencing the 5′-end fragments of the 16S rRNA genes. One sequence was identical to that of the gene of Anaplasma marginale, an etiological agent of animal anaplasmosis. The other sequence was most similar to that of the gene of Ehrlichia chaffeensis, an etiological agent of human monocytic ehrlichiosis. The sequence of 1,501 bases from the novel ehrlichial agent was obtained and showed the greatest levels of sequence similarity (97 to 98%) to 16S rRNA gene sequences of the members of the E. canis group of the genus Ehrlichia. Sequence comparison of the 16S rRNA gene with the members of the genus Ehrlichia reveals that the novel ehrlichial agent detected in B. microplus ticks is a new species of the genus Ehrlichia and is most closely related to E. chaffeensis.
Ehrlichia species are known as important pathogens of medical, as well as veterinary, importance. They are intracellular microorganisms residing within the cytoplasmic vacuoles of monocytes, granulocytes, or platelets of humans and animals. Ehrlichia species elicit illnesses with fever, headache, leukopenia, and thrombocytopenia (9, 13). The genus Ehrlichia is divided into three distinct genogroups based on the similarity of the nucleotide sequence of the 16S rRNA gene: the Ehrlichia canis group (including E. canis, E. chaffeensis, E. muris, and E. ewingii), the E. phagocytophila group (E. phagocytophila, E. equi, human granulocytic ehrlichiosis [HGE] agent, and E. platys), and the E. sennetsu group (E. sennetsu, E. risticii, and the Stellantchasmus falcatus [SF] agent) (2, 15). The members of the E. sennetsu group are transmitted by aquatic vectors (16). In contrast, the members of the E. canis group and the E. phagocytophila group are both tick-borne organisms (9, 15, 16). E. chaffeensis and the HGE agent are tick-borne ehrlichial organisms discovered in the United States. E. chaffeensis, an etiological agent of human monocytic ehrlichiosis, transmitted by the tick Amblyomma americanum, was discovered in 1986, while the HGE agent transmitted by Ixodes scapularis was first reported in 1994 (3, 7-10).
Recently, serological and PCR-based studies suggested that human monocytic ehrlichiosis and HGE also exist outside the United States, particularly in some European and Asian countries (4-7, 11, 14). E. muris, originally isolated from a wild mouse and later isolated from Haemaphysalis flava ticks in Japan, was identified as an agent closely related to E. chaffeensis in serological and genetic analyses (11, 15). Another new Ehrlichia species closely related to E. chaffeensis was isolated from Ixodes ovatus ticks in Japan (14). PCR-based assays and DNA sequencing detected E. chaffeensis, E. canis, and E. platys in various ticks from southern China, while the HGE agent was identified in Ixodes persulcatus ticks from northeastern China (5, 6, 12).
Boophilus microplus ticks are widely distributed over the world and are recognized as the vectors of Anaplasma marginale, an etiological agent of animal anaplasmosis (1). A. marginale is considered to be a member of the tribe Ehrlichieae; 16S rRNA gene analysis shows that it is closely related to the members of the E. phagocytophila group (2, 15). B. microplus ticks are widely distributed in China, where Coxiella burnetii and Borrelia burgdorferi were isolated from the tick species. In this study we used PCR-based assays and DNA sequencing analysis to assess the occurrence of infection by Ehrlichia in B. microplus ticks collected from Sichuan and Tibet, in order to collect data that can be used to provide future estimates of the likelihood of ehrlichiosis in southwestern China.
MATERIALS AND METHODS
Ticks.
B. microplus ticks were collected from infested cattle in Sichuan Province and the Tibet Autonomous Region of China. Immediately after collection, the ticks were immersed in 70% ethanol and stored.
Preparation of DNA extracts from ticks.
Two ticks were placed in a 1.5-ml Eppendorf tube, and then the tube was baked at 100°C for 10 min in an oven. Immediately after baking, the ticks were pulverized in the tube using a glass bar. Following this, 300 μl of Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0), 40 μl of 10% sodium dodecyl sulfate, and 5 μl of proteinase K (20 mg/ml) were added to the tube. The mixture was incubated at 55°C for 3 h and was then incubated in boiling water for 10 min. After a brief centrifugation, the supernatant was transferred to a fresh 1.5-ml tube. The supernatant was extracted twice, first with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and then with an equal volume of chloroform. The aqueous phase (top layer) was transferred to a fresh 1.5-ml tube, the DNA in the aqueous phase was precipitated with ethanol according to the standard method, and the DNA pellets were dissolved in 50 μl of Tris-EDTA buffer and stored at −20°C.
Nested PCR amplification of ehrlichial 16S rRNA genes.
A pair of universal primers, Eh-out1 (5′-TTGAGAGTTTGATCCTGGCTCAGAACG-3′, located at positions 1 to 27 in a 16S rRNA sequence obtained in this study [GenBank accession number AF414399]), and Eh-out2 (5′-CACCTCTACACTAGGAATTCCGCTATC-3′, at positions 653 to 627), were used for the primary amplification. Primers Eh-gs1 (5′-GTAATAACTGTATAATCCCTG-3′, at positions 167 to 187), and Eh-gs2 (5′GTACCGTCATTATCTTCCCTA-3, at positions 448 to 428), which were designed based on the conserved positions in the sequences of 16S rRNA genes of several tick-borne species of Ehrlichia, were used in the nested amplification.
PCR amplifications were performed in 25-μl reaction volumes in a thermal cycler. For the primary amplification, a reaction mixture contained 1.5 pmol each of primers Eh-out1 and Eh-out2, 200 μmol of each deoxynucleoside triphosphate, 2.5 mM MgCl2, 2.5 μg of bovine serum albumin (20 μg/ml), 2.5 μl of 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]), 1 U of Taq DNA polymerase, and 2 μl of tick DNA. The reaction mixture was overlaid with mineral oil and incubated for 5 min at 95°C, followed by 40 cycles of 95°C for 45 s, 55°C for 50 s, and 72°C for 1 min and then a final incubation at 72°C for 5 min to allow complete strand extension.
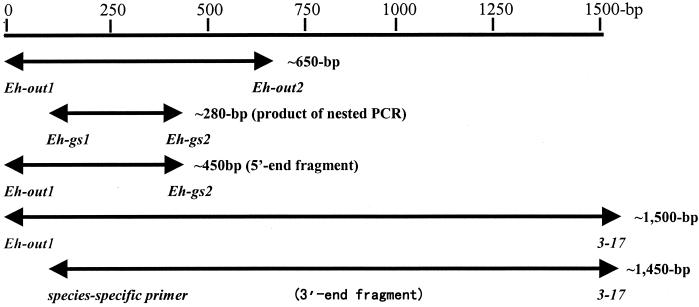
For the nested PCR, the components and conditions were similar to those used in the primary amplification, except that 0.6 μmol of Eh-gs1 and 0.6 μmol of Eh-gs2 were used as primers and that 2 μl of the primary PCR product was used as templates (Fig. 1).
FIG. 1.
The different fragments of 16S rRNA genes amplified with different pairs of primers from the tick DNA specimens. The italicized words represent the names of primers, and the bold lines represent the sizes of the PCR products.
A seminested PCR assay was used to amplify the 5′-end fragments of ehrlichial 16S rRNA genes from the positive specimens of the nested PCR assay. The components and conditions in this assay were similar to that of the nested PCR assay, except that Eh-out1 and Eh-gs2 were used as a pair of seminested primers (Fig. 1).
The 3′-end fragments of ehrlichial 16S rRNA genes were amplified from the positive DNA specimens by using universal primers Eh-out1 and 3-17 (5′-TAAGGTGGTAATCCAGC-3′, at positions 1501 to 1485) in the primary amplification and using a species-specific primer and the 3-17 universal primer in seminested PCR (Fig. 1).
The PCR products were electrophoresed on a 1.5% agarose gel, stained with ethidium bromide, and visualized under UV light. Quality control included both positive and negative controls that were PCR amplified in parallel with all specimens.
Cloning and sequencing of PCR products.
After electrophoresis, the positive DNA fragments were recovered from the agarose gel with a DNA gel extraction kit (NSBC-Sangon, Shanghai, China) according to the instructions of the manufacturer. The fragments were cloned by using the PUCm-T vector (NSBC-Sangon) and competent cells (E. coli DH5α) following the manufacturer's protocol. The recombinant plasmids were extracted from the transformed E. coli cells with a plasmid DNA preparation kit (NSBC-Sangon). Sequencing was carried out by fluorescence-labeled dideoxynucleotide technology with an automated DNA sequencer (SM377; Perkin-Elmer, Norwalk, Conn.). The primers used in the above PCR assays, M13 (5′-CCCAGTCACGACGTTGTAAAACG-3′), located in the PUCm-T vector), and two additional primers (5′-TAGTCCACGCTGTAAACG-3′, at positions 761 to 778; and 5′-CCCGTCAATTCCTTTGAG-3′, at positions 884 to 866), were chosen as sequencing primers.
Computer analyses of DNA sequences.
The analyses of sequence data and sequence alignment were performed on a computer using DNAsis software (Hitachi Software, San Bruno, Calif.). The evolutionary distance calculations and phylogenetic tree inference were performed with the software package CLUSTALX.
Nucleotide sequence accession number.
The GenBank accession numbers for the 16S rRNA genes used for comparison in this study were E. chaffeensis, M73222; E. canis, M73221; E. muris, U15527; E. ewingii, U96436; E. bovis, U03775; E. phagocytophila, M73220; E. platys, AF286699; E. sennetsu, M73225; E. risticii, M21290; and Ehrlichia sp. strain SF, 34280. The nucleotide sequence of the 16S rRNA gene of Ehrlichia sp. strain Tibet was deposited in GenBank under accession number AF414399.
RESULTS
Nested PCR screening of the tick DNA specimens.
A DNA fragment of approximately 280 bp was amplified from 16 of 43 (37%) specimens of B. microplus from Tibet. DNA samples from 27 specimens of B. microplus from Sichuan were all negative. The 280-bp fragments from five of the positive specimens were directly sequenced on the automated DNA sequencer by using Eh-gs1 and Eh-gs2 as sequencing primers. All of the sequences are recognizable as ehrlichial 16S rRNA genes, but the fragment analyzed did not permit them to be distinguished at the species level.
Analyses of the 16S rRNA gene sequences.
In order to identify ehrlichial agents at the species level, the 5′-end fragments (∼450 bp) of 16S ribosomal DNA containing ehrlichial species-specific sequences were amplified from 10 of the nested-PCR-positive specimens by a seminested PCR. The 5′-end fragments were cloned, and the fragments inserted in the recombinant plasmids were sequenced. Four of 10 sequences were identical to a 16S rRNA gene sequence of A. marginale (GenBank accession number AF311303). The remaining 6 of 10 sequences were most similar to a 16S rRNA gene sequence of E. chaffeensis (Arkansas strain; GenBank accession number M73222).
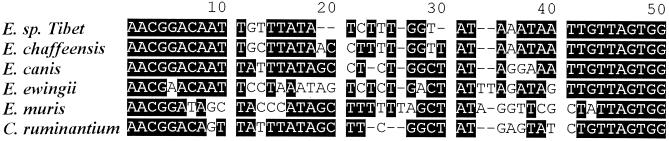
A fragment of approximately 1,450 bp was amplified by a seminested PCR from the samples that were shown to contain the novel ehrlichial agent as demonstrated by sequences from the 5′-end fragments. This seminested PCR used primer pair Eh-out1 and 3-17 together with another pair of primers (universal primer 3-17 and a species-specific primer [5′-CGAACGGACAATTGTTTATATC-3′] designed based on the highly variable region of the 16S rRNA gene of the novel agent [Fig. 2 ]). A sequence of 1,501 bases was obtained by linking the 5′-end and 3′-end sequences of the 16S rRNA gene of the novel agent, based on overlapping regions of sequence. When the entire sequence of the 16S rRNA gene of the novel agent is compared to those of other species of Ehrlichia, the greatest levels of nucleotide sequence similarity (97 to 98%) were found with the members of the E. canis group of the genus Ehrlichia. The entire sequence was most similar to the 16S rRNA gene sequences of E. chaffeensis but differed by 19 nucleotides (∼1.4%).
FIG. 2.
A highly variable region of sequence located at the 5′ end of the 16S rRNA gene revealed by multiple alignment of 16S rRNA gene sequences of Ehrlichia spp. in the E. canis genogroup. C. ruminantium, Cowdria ruminantium.
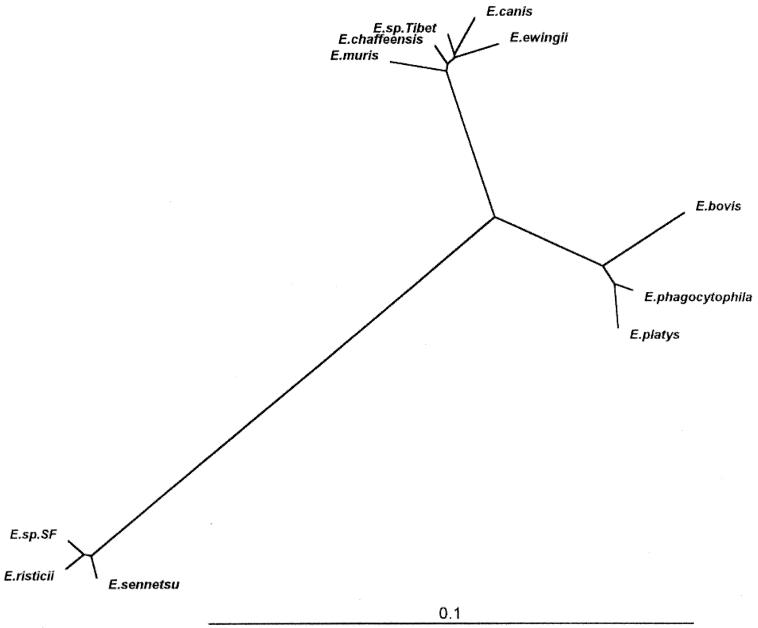
The entire sequence of the novel agent was aligned with the 16S rRNA gene sequences of the known species of the genus Ehrlichia by using a multiple sequence alignment program of DNAsis, and a 1,403-base portion of the 16S rRNA gene sequence that could be unambiguously aligned for all species of the genus Ehrlichia was used for phylogenetic analysis. The levels of sequence divergences and similarities between the novel agent and the organisms of the genus Ehrlichia are shown in Table 1. Consistent with the results above, the greatest levels of similarity (98 to 99%) for the 1,403-bp region were found between the novel agent and the sequences of the E. canis group; the highest level of similarity (99%) was found between it and the sequence of E. chaffeensis (Table 1). The phylogenetic tree obtained from the data is shown in Fig. 3. It is clear that the novel ehrlichial agent in the ticks from Tibet belongs to the E. canis group within the genus Ehrlichia and was not closely related to the E. phagocytophila or E. sennetsu genogroup (Table 1 and Fig. 3). The analyses of the levels of sequence divergence (Table 1) show that divergence between the novel agent and its closest neighbor, E. chaffeensis, is sometimes greater than the levels of sequence divergence obtained in pairwise comparisons of several other Ehrlichia species.
TABLE 1.
Levels of genetic similarity and differences between novel agent (Tibet strain) and ehrlichial species in 16S rRNA sequencesa
| Organism | % Similarity or difference between Ehrlichia sp. strain Tibet and:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ehrlichial sp. strain Tibet | E. chaffeensis | E. canis | E. ewingii | E. muris | E. phagocytophila | E. platys | E. bovis | E. risticii | E. sennetsu | Ehrlichia sp. strain SF | |
| Ehrlichia sp. strain Tibet | 0.010 | 0.013 | 0.014 | 0.021 | 0.072 | 0.080 | 0.087 | 0.157 | 0.154 | 0.154 | |
| E. chaffeensis | 99.00 | 0.016 | 0.016 | 0.016 | 0.071 | 0.078 | 0.084 | 0.154 | 0.152 | 0.152 | |
| E. canis | 98.72 | 98.43 | 0.018 | 0.027 | 0.074 | 0.081 | 0.088 | 0.157 | 0.155 | 0.155 | |
| E. ewingii | 98.65 | 98.36 | 98.22 | 0.021 | 0.076 | 0.083 | 0.091 | 0.158 | 0.157 | 0.155 | |
| E. muris | 97.93 | 98.36 | 97.29 | 97.86 | 0.076 | 0.082 | 0.088 | 0.159 | 0.157 | 0.156 | |
| E. phagocytophila | 92.80 | 92.94 | 92.59 | 92.44 | 92.37 | 0.014 | 0.031 | 0.150 | 0.149 | 0.151 | |
| E. platys | 92.02 | 92.24 | 91.87 | 91.66 | 91.81 | 98.65 | 0.031 | 0.154 | 0.152 | 0.155 | |
| E. bovis | 91.31 | 91.60 | 91.23 | 90.95 | 91.24 | 96.94 | 96.87 | 0.159 | 0.157 | 0.162 | |
| E. risticii | 84.27 | 84.56 | 84.33 | 84.19 | 84.13 | 85.00 | 84.58 | 84.08 | 0.009 | 0.009 | |
| E. sennetsu | 84.56 | 84.85 | 84.48 | 84.33 | 84.35 | 85.14 | 84.48 | 84.30 | 99.07 | 0.013 | |
| Ehrlichia sp. strain SF | 84.56 | 84.85 | 84.55 | 84.55 | 84.42 | 84.93 | 84.51 | 83.80 | 99.07 | 98.72 | |
The values on the upper right are corrected levels of nucleotide differences for 1,403 bases, which could be aligned without ambiguity in all organisms. The values on the lower left are percentages of sequence similarity for regions that could be aligned unambiguously.
FIG. 3.
Phylogenetic relationships between Ehrlichia sp. strain Tibet and other members of the genus Ehrlichia. The tree was constructed using the neighbor-joining program in the software package CLUSTALX.
DISCUSSION
Ehrlichia spp. are obligatory intracellular pathogens. Isolation of Ehrlichia from cell culture is a classical method for determining their presence in blood and tissue specimens of human or animals. PCR is a sensitive and simple method used to directly detect infectious agents in various specimens, and nested PCR is more sensitive than standard PCR procedures (17). A nested PCR assay specific for amplification of the 16S rRNA genes of tick-borne Ehrlichia spp. was established using nested 16S rRNA primers specific only for the tick-borne Ehrlichia species. Using this nested PCR analysis, 37% of DNA samples of ticks from Tibet were positive, while the DNA samples of ticks from Sichuan were all negative. Further, sequence analysis of the amplimers showed that they were recognizable as fragments of 16S rRNA genes of tick-borne Ehrlichia. These results suggest that the nested PCR is both highly sensitive and specific for detection of the tick-borne Ehrlichia spp., and it is particularly useful for screening the specimens containing the new species of the genus Ehrlichia.
Since the 280-bp amplimers produced by nested PCR did not contain the species-specific signature sequences of Ehrlichia spp., the ehrlichial agents in the positive specimens could not be identified at the species level by sequencing these amplimers. Therefore, the 5′-end fragments of 16S rRNA genes of the ehrlichial agents in the positive specimens were amplified and sequenced. The analyses of 5′-end fragments revealed that the positive specimens contained two agents: A. marginale, an etiological agent of animal anaplasmosis, and a novel ehrlichial agent that was most closely related to E. chaffeensis, an etiological agent of human monocytic ehrlichiosis. However, the question of whether the two agents coexist in B. microplus remains to be studied, because each DNA sample in this study was prepared from two ticks.
Sequence comparison of the 16S rRNA gene is recognized as one of the most powerful and precise methods for determining the phylogenetic relationships of bacteria (2, 15, 16, 18). In this study, our analysis of 16S rRNA sequences revealed that the novel agent found in the Tibet ticks was a member of the E. canis group of the genus Ehrlichia and was most closely related to E. chaffeensis but was also closely related to E. canis, E. ewingii, and E. muris. Since the 16S rRNA gene is known to exhibit a high level of structural conservation and a low evolutionary rate (18), levels of sequence divergence greater than 0.5% in comparisons between nearly complete 16S rRNA gene sequences of members of the genus Ehrlichia have been considered sufficient to classify organisms as different species (2, 15, 16). The levels of divergence of the 16S rRNA sequence between the novel Tibetan ehrlichial agent and the members of the E. canis group were approximately 1 or 2% in pairwise comparisons of 1,403-base sequences, and this level of difference should be sufficient to classify the novel ehrlichial agent as a new species of the genus Ehrlichia. Since the novel agent was first detected in the ticks from Tibet, it is temporarily named Ehrlichia sp. strain Tibet.
Although it is well known that A. marginale is an etiological agent of bovine anaplasmosis and is transmitted by B. microplus ticks (1), the detection of any species of Ehrlichia in B. microplus ticks has not been reported before. The identification of A. marginale and Ehrlichia sp. strain Tibet in B. microplus ticks suggests the possibility of coinfection and cotransmission of the two agents in the area where the ticks were collected. Therefore, it is also possible that, in this area, B. microplus ticks and the cattle infested by these ticks are coinfected with the two agents. However, these probabilities remain to be demonstrated by further studies.
Acknowledgments
We are grateful to Paul A. Fuerst (Department of Evolution, Ecology and Organismal Biology, Department of Molecular Genetics, The Ohio State University, Columbus) for critical comments on the manuscript.
This study was supported by a grant from National Natural Science Foundation of China (No. 39870042).
REFERENCES
- 1.Aguirre, D. H., A. B. Gaido, A. E. Vinabal, S. T. De Echaide, and A. A. Guglielmone. 1994. Transmission of Anaplasma marginale with adult Boophilus microplus ticks fed as nymphs on calves with different levels of rickettsaemia. Parasite 1:405-407. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B. E., K. G. Simms, J. G. Olson, J. E. Childs, J. F. Piesman, C. M. Happ, G. O. Maupin, and B. J. Johnson. 1993. Amblyomma americanum: a potential vector of human ehrlichiosis. Am. J. Trop. Med. Hyg. 49:239-244. [DOI] [PubMed] [Google Scholar]
- 4.Baumgarten, B. U., M. Röllinghoff, and C. Bogdan. 1999. Prevalence of Borrelia burgdorferi and granulocytic and monocytic ehrlichiae in Ixodes ricinus ticks from southern Germany. J. Clin. Microbiol. 37:3448-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, W.-C., Q.-M. Zhao, P.-H. Zhang, J. S. Dumler, X.-T. Zhang, L.-Q. Fang, and H. Yang. 2000. Granulocytic ehrlichiae in Ixodes persulcatus ticks from an area in China where Lyme disease is endemic. J. Clin. Microbiol. 38:4208-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, W.-C., Y.-M. Cao, P.-H. Zhang, X.-T. Zhang, Q.-H. Dai, J. S. Dumler, L.-Q. Fang, and H. Yang. 2000. Identification of Ehrlichia chaffeensis by nested PCR in ticks from southern China. J. Clin. Microbiol. 38:2778-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. Walker. 1994. Identification of a granulocytic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:584-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumler, J. E., and J. S. Bakken. 1995. Ehrlichial diseases of humans: emerging tick-borne infections. Clin. Infect. Dis. 20:1102-1110. [DOI] [PubMed] [Google Scholar]
- 10.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 11.Kawahara, M., T. Ito, C. Suto, S. Shibata, Y. Rikihisa, K. Hata, and K. Hirai. 1999. Comparison of Ehrlichia muris strains isolated from wild mice and ticks and serologic survey of humans and animals with E. muris as antigen. J. Clin. Microbiol. 37:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan, H., Y. H. Ma, S. D. Tang, Y. Sun, B. Wen, and X. R. Chen. 2000. Canine ehrlichiosis caused simultaneously by Ehrlichia canis and Ehrlichia platys. Microbiol. Immunol. 44:737-739. [DOI] [PubMed] [Google Scholar]
- 13.Rikihisa, Y. 1991. The tribe Ehrlichieae and ehrlichial diseases. Clin. Microbiol. Rev. 4:286-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata, S.-H., M. Kawahara, Y. Rikihisa, H. Fujita, Y. Watanaba, C. Suto, and T. Ito. 2000. New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J. Clin. Microbiol. 38:1331-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen, B., Y. Rikihisa, J. Mott, P. A. Fuerst, M. Kawahara, and C. Suto. 1995. Ehrlichia muris sp. nov., identified on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int. J. Syst. Bacteriol. 45:250-254. [DOI] [PubMed] [Google Scholar]
- 16.Wen, B., Y. Rikihisa, S. Yamamoto, N. Kawabata, and P. A. Fuerst. 1996. Characterization of the SF agent, an Ehrlichia sp. isolated from the fluke Stellantchasmus falcatus, by 16S rRNA base sequence, serological, and morphological analyses. Int. J. Syst. Bacteriol. 46:149-154. [DOI] [PubMed] [Google Scholar]
- 17.Wen, B., Y. Rikihisa, J. M. Mott, R. Greene, H.-Y. Kim, N. Zhi, G. C. Couto, A. Unver, and R. Bartsch. 1997. Comparison of nested PCR with immunofluorescent-antibody assay for detection of Ehrlichia canis infection in dogs treated with doxycycline. J. Clin. Microbiol. 35:1852-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]