Abstract
Instability and plasticity of telomeric DNA, which includes extrachromosomal DNA, are usually correlated with the absence of telomerase and with abnormal growth of mammalian cells. Here, we show the formation of extrachromosomal circular DNA of telomeric repeats (tel-eccDNA) during the development of Xenopus laevis. Tel-eccDNA is double-stranded relaxed circles composed of the vertebrate consensus telomeric repeats (TTAGGG)n. Its size varies from <2 to >20 kb and it comprises up to 10% of the total cellular telomere content of the early embryo (pre-MBT stage). The amount of tel-eccDNA is reduced in later developmental stages and in adult tissues. Using a cell-free system derived from Xenopus egg extracts, we show that tel-eccDNA can be formed de novo from the telomere chromosomal tracts of sperm nuclei and naked DNA in a replication-independent manner. These results reveal an unusual plasticity of telomeric DNA during normal development of Xenopus.
Introduction
Telomeres are structurally and functionally specialized DNA–protein complexes at the end of eukaryotic chromosomes. They play an important role in ensuring genome integrity by protecting the chromosome ends from degradation by nucleases and from chromosome fusion and provide the structural basis for solving the end-replication problem. In the absence of telomerase, telomeric DNA may be involved in processes typical of genomic instability, such as hyper-recombination, amplification and rapid deletion (Niida et al., 2000; Lundblad, 2002).
One example of genome plasticity and instability is the formation of extrachromosomal circular DNA (eccDNA), also termed small polydispersed circular DNA (spcDNA), presumably derived from repetitive chromosomal sequences. Amongst the sequences identified in the population of eccDNA were direct tandem repeats of both coding and non-coding chromosomal sequences (Pont et al., 1987; Cohen et al., 1999), organized as multimers of the repeating units. EccDNA is thought to emerge from the chromosomes by a not-fully-understood mechanism that may involve homologous recombination (Cohen and Méchali, 2001; reviewed in Gaubatz, 1990). EccDNA consisting of sequences identical to telomeric repeats was detected in rodent cells and in some human tumors and cell lines (Regev et al., 1998). Recently, circles that consist of multimers of telomeric repeat sequences of the linear mitochondrial genome of the yeast Candida parapsilosis were also reported (Tomaska et al., 2000). Extrachromosomal circles of telomeric DNA of higher eukaryotes have been observed in cancer cells but not in normal cells. Therefore, it is important to determine whether they occur during the normal life-cycle of vertebrates or whether they represent cellular phenomena peculiar to derailed development or are specific to special organelles of lower eukaryotes.
In Xenopus laevis, the ends of the telomeres are composed of long tracts of the vertebrate consensus telomeric repeat (TTAGGG)n, and their size ranges from <10 to >48 kb (Bassham et al., 1998). We have reported previously a developmentally regulated formation of eccDNA that occurs in pre-blastula Xenopus and can be reproduced in Xenopus egg extracts (Cohen et al., 1999; Cohen and Méchali, 2001). Multimers of tandemly repeated sequences were over-represented in the circle population, whereas dispersed sequences were not detected, indicating a non-random mechanism of circle formation. In this work, we report the presence of high levels of telomeric eccDNA (tel-eccDNA) in early Xenopus embryos. This population of circles is less abundant in later developmental stages and in adult tissues. We show that the production of tel-eccDNA can be reproduced in vitro in Xenopus egg extracts when sperm nuclei or naked DNA serve as substrates. Very recent observations in yeast suggest that artificial circular DNA molecules might generate recombinational telomere elongation (Natarajan and McEachern, 2002). The present paper emphasizes these data by showing that such circles do exist in vivo during the development of a vertebrate.
Results
Extrachromosomal circular telomeric DNA is present in early Xenopus embryos
We have previously demonstrated the developmentally regulated formation of eccDNA molecules in early Xenopus embryos (Cohen et al., 1999). To determine whether the telomeric repeats of Xenopus were represented in vivo in the eccDNA population, genomic DNA from early embryos was separated on a two-dimensional (2D) gel and hybridized with the oligonucleotide (TTAGGG)5 as a probe. We detected in the early embryos an abundant population of relaxed circles homologous to the telomeric sequence (tel-eccDNA), comprising up to 10% of the total cellular telomeric DNA content (Figure 1B). To determine the size and structure of these telomere-homologous circles, we used circular plasmids relaxed by topoisomerase I, loaded onto the gel together with the embryonic DNA (Figure 1C). Tel-eccDNA co-migrated with the relaxed plasmids (Figure 1D) and was heterogeneous in mass, ranging from 2 to >22 kb, indicating that the length of tel-eccDNA can be as large as an entire chromosomal telomere tract.
Figure 1.
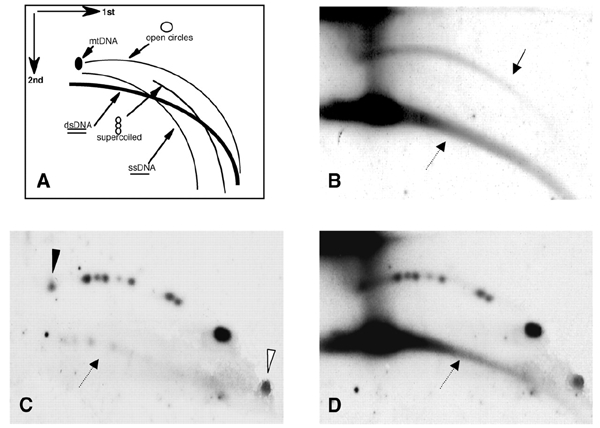
Tel-eccDNA is detected in Xenopus early embryos. (A) 2D gel electrophoretic patterns of genomic DNA generated by populations of linear and circular molecules (Cohen and Lavi, 1996). Each arc consists of molecules sharing the same structure but differing in mass. dsDNA, doublestranded DNA; ssDNA, single-stranded DNA; mtDNA, mitochondrial DNA; open circles, circular double-stranded DNA; supercoiled, double-stranded supercoiled DNA. (B–D) DNA from Xenopus early blastula (2000-cell stage), mixed with pUC18-derived plasmid relaxed-circle size markers (see text) and separated on a 2D gel. The blot was hybridized with telomeric repeat probe (TTAGGG)5 (B) and then with the plasmid probe (pUC18), to identify relaxed circles (C). The plasmids range from 2.7 kb (open arrowhead) to 22.4 kb (solid arrowhead). The solid arrow points to the relaxed circles, and the dashed arrows point to the linear DNA. (D) Co-migration of the telomere-homologous DNA arc with the circular markers is demonstrated by the superposition of (B) and (C).
Tel-eccDNA is double-stranded
T-loops have been reported as the circular structure of the end of telomeres in mammalian cells, held by proteins (Griffith et al., 1999; Stansel et al., 2001). One could propose that tel-eccDNA originated from such T-loops. However, we detected tel-eccDNA after extensive proteinase K digestion and phenol extractions, indicating that, unlike T-loops, DNA ends are not held together by proteins. In addition, preheating of the DNA to 68°C prior to electrophoresis did not affect the circular DNA, indicating that these molecules were not closed by a weak annealing between short cohesive ends (data not shown).
The co-migration of tel-eccDNA with relaxed plasmids suggests that it consists of double-stranded DNA. To further establish this point, we treated the embryonic DNA with S1 nuclease prior to its 2D-gel analysis (Figure 2). Singlestranded bacteriophage φX174 DNA was mixed with the embryonic DNA as an internal control. Figure 2 shows that tel-eccDNA was resistant to S1 nuclease, and no change in its amount or structure was observed after 30 min of incubation with S1 (Figure 2A–C), although 5 min of incubation was enough to digest most of the φX174 DNA (Figure 2E). We conclude that tel-eccDNA is entirely doublestranded.
Figure 2.
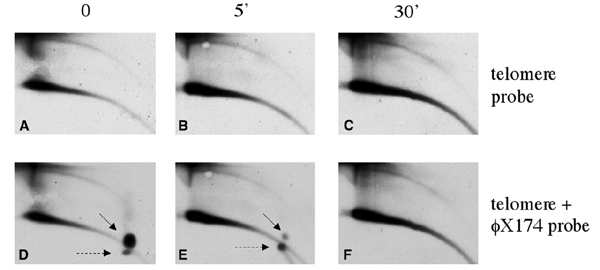
Tel-eccDNA is resistant to S1 nuclease. Total genomic DNA from Xenopus early embryos was mixed with 1 μg of φX174 single-stranded DNA and treated with S1 nuclease. A sample was taken before adding the enzyme (A and D), and the other samples were taken after 5 min (B and E) and 30 min (C and F) of incubation. DNA was analyzed on a 2D gel and hybridized with telomeric probe (A–C) and φX174 probe (D–E) to follow degradation of single-stranded DNA. The solid arrows point to the single-stranded circular form of φX174, and the dashed arrows point to its linear form.
Tel-eccDNA consists mostly, if not exclusively, of telomeric repeats
To determine whether tel-eccDNA molecules consist of telomeric repeats or contain additional genomic sequences, embryonic DNA was digested with the frequent cutters MspI and CfoI, which leave the telomeric repeats intact but cleave the genomic DNA. Upon hybridization with the telomere probe, we observed no difference in the amount or size range of the circular telomere-homologous molecules between cut and uncut DNA (Figure 3A and B). In contrast, the circular multimers of satellite 1 (Figure 3D), a non-telomeric 741–750 bp unit, organized in tandem in long chromosomal tracts, which is over-represented in the eccDNA population (Cohen et al., 1999), were completely degraded (Figure 3C). Ethidium-bromide staining of the gel indicated digestion of the total genomic DNA, and we also observed that tel-eccDNA was resistant to cleavage by other frequently cutting enzymes such as AluI, RsaI and DraI (data not shown). These observations indicate that tel-eccDNA molecules do not contain sequences that are sensitive to frequent cutters and therefore suggest that they mostly, if not exclusively, consist of telomeric repeats.
Figure 3.
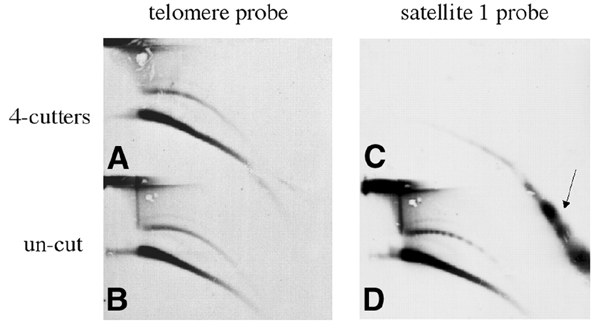
Tel-eccDNA consists exclusively of telomeric repeats. Genomic DNA from Xenopus early embryos was digested with the frequent cutting enzymes MspI and CfoI (A and C) and analyzed by a 2D gel in comparison with uncut DNA (B and D). The second dimension was carried out by running the two lanes corresponding to the first dimension on the top and bottom of the same agarose gel. Tel-eccDNA was not affected by the enzymatic cleavage (A and B), whereas satellite 1 (Lam and Carroll, 1983) was extensively cleaved as the whole bulk of genomic DNA. In the uncut DNA (D), a ladder of circular multimers of the satellite 1 unit (multimers of 741–750 bp) is detected. The arrow points to the low-molecular-weight linear fragments of satellite 1 following the MspI–CfoI cleavage.
The amount of tel-eccDNA is under developmental regulation
The amount of tel-eccDNA relative to telomeric genomic DNA (double-stranded DNA arc) decreases after mid-blastula transition (MBT, Figure 4), as observed previously for genomic and satellite I DNA (Cohen et al., 1999; Figure 4). However, we reproducibly observed that tel-eccDNA is still detected at late developmental stages when other sequences are not, even when increased amounts of total DNA and over-exposures of 2D gels are performed (Figure 4). We estimated a 3-fold relative decrease of tel-eccDNA at the neurula stage, as opposed to at least a 50-fold decrease for satellite I or other chromosomal sequences (Figure 4; data not shown). We conclude that the population of tel-eccDNA decreases during late development but that a fraction is still present when other ecc sequences are not.
Figure 4.
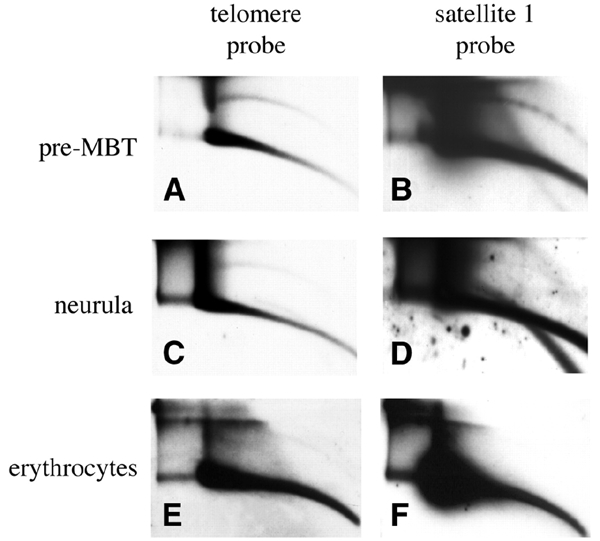
The level of tel-eccDNA is decreased during development. Genomic DNA from Xenopus embryos and Xenopus erythrocytes was separated on a 2D gel: (A and B) 590 ng of early blastula embryos (pre-MBT, 1000-cell stage); (C and D) 1.4 μg of late neurula (90000 cells per embryo); (E and F) 1 μg of erythrocytes DNA. The quantity of embryonic DNA was estimated from the embryonic stage and checked by hybridization with a genomic DNA probe in comparison with DNA calibrated at 260 nm and loaded on the same gel (data not shown). The blots were hybridized with telomeric (A, C and E) and satellite 1 (B, D and F) probes.
A Xenopus cell-free system produces tel-eccDNA de novo
An important question is the relation between extrachromosomal circular forms and their chromosomal counterparts (Regev et al., 1998; Tomaska et al., 2000). We have previously shown that activated egg extracts of Xenopus are capable of forming eccDNA de novo when incubated with sperm nuclei or naked DNA (Cohen et al., 1999; Cohen and Méchali, 2001). We specifically tested here whether tel-eccDNA can be generated in vitro by a similar mechanism. Following incubation of sperm nuclei in egg extract, an arc of tel-eccDNA appeared, comprising ∼10% of the total telomeric DNA content (Figure 5A). This level is identical to the tel-eccDNA levels observed in early embryos (Figures 1 and 4). Tel-eccDNA was not detected in sperm DNA (Figure 5G) or in the egg-extract, since its incubation without template did not result in tel-eccDNA (data not shown). We conclude that tel-eccDNA is formed de novo from the chromosomes.
Figure 5.
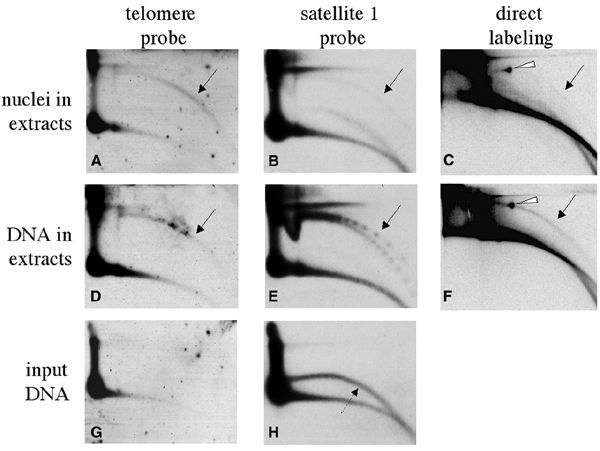
In vitro formation of tel-eccDNA from sperm nuclei and from naked sperm DNA. Sperm nuclei (270 ng of DNA) (A–C) or 300 ng of purified naked sperm DNA (D–F) were incubated for 2 h in 100 μl of low-speed Xenopus egg extract in the presence (C and F) or absence (A, B, D and E) of 50 μCi of [α-32P]dCTP. Following incubation, the DNA was extracted and analyzed by a 2D gel as well as non-incubated naked sperm DNA (G and H). The gel was hybridized with telomeric probe (A, D and G) and with satellite 1 probe (B, E and H). The gel that contained the radiolabeled samples was exposed to autoradiography (C and F). The solid arrows point to arcs of relaxed circles. In (E), a series of supercoiled multimers is also identified. The dashed arrow points to single-stranded DNA in the preparation of naked sperm DNA, which was previously shown to be irrelevant to eccDNA formation, as its degradation by S1 nuclease does not alter the production of eccDNA (Cohen and Méchali, 2001). The white arrowheads point to mtDNA that is present in the extract and was also labeled.
Tel-eccDNA is formed from naked DNA at a similar level as from sperm nuclei (Figure 5D). In contrast, eccDNA of non-telomeric sequences is formed in vitro much less efficiently from sperm nuclei than from naked DNA, as detected by re-hybridization with satellite 1 (Figure 5B and E) and in experiments of direct labeling (Figure 5C and F). These results suggest that telomeric regions in sperm chromatin are more accessible to the proteins involved in the formation of extrachromosomal circles than other non-telomeric sequences.
Formation of tel-eccDNA in vitro begins prior to, and occurs independently of, chromosomal DNA replication
De-membraned sperm nuclei replicate with a lag of 40 min (Blow and Laskey, 1986; Menut et al., 1999; Figure 6A). Figure 6B shows that an arc of tel-eccDNA is detected after only 5 min of incubation, at a stage when sperm chromatin is still condensed and nuclear membrane has not yet formed (data not shown). After 30 min of incubation, before the onset of chromosomal DNA replication, it comprised ∼7% of the total telomere content. When replication of the chromosomal DNA is completed (Figure 6B, 120′), the relative amount of tel-eccDNA remained stable, varying between 8 and 11% of the total telomere content in different experiments (Figure 6; data not shown). This result suggests that the formation of tel-eccDNA occurs independently of chromosomal DNA replication. This is in agreement with the observation that the addition of aphidicolin (an inhibitor of DNA polymerase) at the beginning of the reaction did not affect the formation of tel-eccDNA tested after 30 min (before the onset of replication; data not shown) or 120 min of incubation (Figure B, 120′ aphi).
Figure 6.
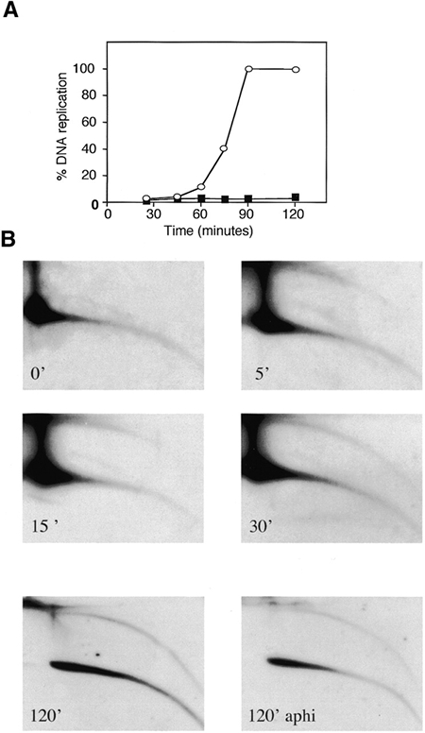
Kinetics of tel-eccDNA formation in vitro. (A) Sperm nuclei were incubated in egg extracts in the presence (open circles) or absence (solid squares) of 100 μg/ml aphidicolin. DNA replication kinetics was followed during a 2 h period by incorporation of [α-32P]dCTP. Samples were taken at the indicated times for 2D-gel analysis followed by hybridization with telomere probe (B).
Discussion
We describe the presence of eccDNA molecules consisting of the consensus telomere repeats in Xenopus. These are double-stranded relaxed circles and are stable, as protease-resistant DNA, suggesting that they are not T-loops (Griffith et al., 1999). The size of tel-eccDNA varies from <2 to >22 kb, indicating that large parts of the chromosomal telomeres may be mobilized.
Telomere plasticity and mobility in cells is usually correlated with the absence of telomerase (Lundblad, 2002). Extrachromosomal telomeric DNA was observed in telomerase-negative immortalized human cells (Tokutake et al., 1998) and was proposed to be involved in the mechanism of telomere maintenance. It was also detected in ATM−/− mice and in patients with ataxia-telangiectasia (Hande et al., 2001). We found tel-eccDNA during the normal development of Xenopus embryos, when telomerase activity is still detected (Mantell and Greider, 1994). Tel-eccDNA is highly enriched in early embryonic development (pre-MBT) of Xenopus and may comprise 10% of the total content of the cellular telomeric repeats. This population decreases in later developmental stages, but tel-eccDNA can still be detected in late development stages when other ecc sequences were not, suggesting a high plasticity of telomeric sequences.
Tel-eccDNA was previously observed in rodent and human cells (Regev et al., 1998) and believed to originate from the chromosomal telomeric repeats, although no direct evidence was presented. In the present work, using the Xenopus cell-free system, we show that de novo formation of tel-eccDNA requires Xenopus chromatin or DNA of sperm nuclei. Formation of tel-eccDNA occurs rapidly, before the onset of chromosomal DNA replication. The possibility that tel-eccDNA was artifactually formed by self-ligation of fragmented chromosomes is unlikely, since linear DNA with the vertebrate telomere (TTAGGG)n ends was protected from ligation in vitro in Xenopus egg extracts and in vivo when injected to Xenopus eggs (Li et al., 1998).
Tel-eccDNA is composed of telomeric sequences, consistent with observations in mammalian cells (Regev et al., 1998), where no other chromosomal sequences are combined into the array of the telomeric repeats. This result is also consistent with previous reports that the size of eccDNA in Xenopus, yeast, Drosophila and humans corresponds to integral numbers of their unit repeats (Kiyama et al., 1986, 1987; Pont et al., 1987; Sinclair and Guarente, 1997; Cohen et al., 1999; Cohen and Méchali, 2001). Together, these data support a mechanism by which tel-eccDNA (like other tandemly repeated units) is formed by an intrachromosomal homologous recombination between tandem telomeric repeats. As a result, tracts of various sizes can 'loop-out' and form the circles. We suggest that this replication-independent formation of circular DNA molecules may account for the excised telomeric DNA that was proposed as a potential product of telomere rapid deletion in yeast (Bucholc et al., 2001) and for the extrachromosomal circles that contain the Y′ sub-telomeric element reported in Saccharomyces cerevisiae (Horowitz and Haber, 1985). A similar mechanism was described for the formation of extrachromosomal ribosomal circles in S. cerevisiae (Kaeberlein et al., 1999; Park et al., 1999).
We conclude that tel-eccDNA is a common phenomenon that occurs in various eukaryotic organisms from yeast to mammalian cells. In two examples (in Xenopus and in the mitochondria of C. parapsilosis), tel-eccDNA is found during the normal life-cycle and is not confined to malignancy or other abnormal states. In Xenopus, unlike other examples from yeast and cultured cells, tel-eccDNA occurs even at a stage when telomerase activity is still detected (Mantell and Greider, 1994). Whether tel-eccDNA can re-integrate into the chromosomes is an intriguing possibility in the light of models for telomerase-independent mechanisms for telomere maintenance (Horowitz and Haber, 1985; Natarajan and McEachern, 2002).
Methods
Xenopus egg extracts.
Preparation of low-speed egg extracts was performed as described previously (Menut et al., 1999).
Preparation of DNA.
Genomic DNA was prepared by rapidly homogenizing Xenopus embryos or tissues in 30 mM EDTA, 1% SDS, 0.5% Triton X-100 and 0.3 M NaCl, and then incubating at 37°C for 16 h with 0.1 mg/ml proteinase K. DNA was extracted with phenol–chloroform, precipitated with ethanol and digested with 0.2 mg/ml RNase A for 2 h at 37°C.
Neutral–neutral 2D electrophoresis.
Separation of DNA in neutral–neutral 2D gels was performed according to Brewer and Fangman (1987), with modifications as described by Cohen and Lavi (1996). The DNA was first separated on 0.4% agarose at low voltage in 1× TBE, and the gel was stained with 0.3 μg/ml ethidium bromide. The lane was cut and placed on a clean gel support at 90° to the direction of the first electrophoresis, cast with 1.1% agarose containing 0.3 μg/ml ethidium bromide and run in 1× TBE in the presence of 0.3 μg/ml ethidium bromide. The first dimension was run overnight at 1 V/cm, and the second at 4 V/cm for 5 h, both at room temperature. Southern blot analyses were performed with Hybond N+ nylon membranes (Amersham, UK). As a probe for telomeric DNA, we used an end-labeled oligonucleotide (TTAGGG)5. Other probes were labeled by the ready-to-go kit (Pharmacia).
Acknowledgments
We thank D. Segal for critical reading of this manuscript. This study has been supported by grants from the CNRS, Association pour la Recherche sur le Cancer, and the Fondation pour la Recherche Médicale (FRM). S.C. was supported by post-doctoral fellowships from EMBO and FRM.
References
- Bassham S., Beam A. and Shampay J. (1998) Telomere variation in Xenopus laevis. Mol. Cell. Biol., 18, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J. and Laskey R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Bucholc M., Park Y. and Lustig A.J. (2001) Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. and Lavi S. (1996) Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol. Cell. Biol., 16, 2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. and Méchali M. (2001) A novel cell-free system reveals a mechanism of circular DNA formation from tandem repeats. Nucleic Acids Res., 29, 2542–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Menut S. and Méchali M. (1999) Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol. Cell. Biol., 19, 6682–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz J.W. (1990) Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res., 237, 271–292. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H. and de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Hande M.P., Balajee A.S., Tchirkov A., Wynshaw-Boris A. and Lansdorp P.M. (2001) Extra-chromosomal telomeric DNA in cells from Atm−/− mice and patients with ataxia-telangiectasia. Hum. Mol. Genet., 10, 519–528. [DOI] [PubMed] [Google Scholar]
- Horowitz H. and Haber J.E. (1985) Identification of autonomously replicating circular subtelomeric Y′ elements in Saccharomyces cerevisiae. Mol. Cell. Biol., 5, 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M. and Guarente L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev., 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama R., Matsui H. and Oishi M. (1986) A repetitive DNA family (Sau3A family) in human chromosomes: extrachromosomal DNA and DNA polymorphism. Proc. Natl Acad. Sci. USA, 83, 4665–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama R., Matsui H., Okumura K. and Oishi M. (1987) A group of repetitive human DNA families that is characterized by extrachromosomal oligomers and restriction-fragment length polymorphism. J. Mol. Biol., 193, 591–597. [DOI] [PubMed] [Google Scholar]
- Lam B.S. and Carroll D. (1983) Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J. Mol. Biol., 165, 567–585. [DOI] [PubMed] [Google Scholar]
- Li L., Lejnine S., Makarov V. and Langmore J.P. (1998) In vitro and in vivo reconstitution and stability of vertebrate chromosome ends. Nucleic Acids Res., 26, 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. (2002) Telomere maintenance without telomerase. Oncogene, 21, 522–531. [DOI] [PubMed] [Google Scholar]
- Mantell L.L. and Greider C.W. (1994) Telomerase activity in germline and embryonic cells of Xenopus. EMBO J., 13, 3211–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menut S., Lemaitre J.M., L., Hair A. and Méchali M. (1999) DNA replication and chromatin assembly using Xenopus eggs or embryos. In Richter, J.D. (ed.), Advances in Molecular Biology: A Comparative Methods Approach to the Study of Oocytes and Embryos. Oxford University Press, Oxford, UK, pp. 198–226. [Google Scholar]
- Natarajan S. and McEachern M.J. (2002) Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol., 22, 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H., Shinkai Y., Hande M.P., Matsumoto T., Takehara S., Tachibana M., Oshimura M., Lansdorp P.M. and Furuichi Y. (2000) Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol. Cell. Biol., 20, 4115–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P.U., Defossez P.A. and Guarente L. (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont G., Degroote F. and Picard G. (1987) Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J. Mol. Biol., 195, 447–451. [DOI] [PubMed] [Google Scholar]
- Regev A., Cohen S., Cohen E., Bar-Am I. and Lavi S. (1998) Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene, 17, 3455–3461. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Stansel R.M., de Lange T. and Griffith J.D. (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J., 20, 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutake Y. et al. (1998) Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun., 247, 765–772. [DOI] [PubMed] [Google Scholar]
- Tomaska L., Nosek J., Makhov A.M., Pastorakova A. and Griffith J.D. (2000) Extragenomic doublestranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res., 28, 4479–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]


