Abstract
A family of genetically and structurally homologous complexes, the proteasome lid, Cop9 signalosome (CSN) and eukaryotic translation initiation factor 3, mediate different regulatory pathways. The CSN functions in numerous eukaryotes as a regulator of development and signaling, yet until now no evidence for a complex has been found in Saccharomyces cerevisiae. We identified a group of proteins, including a homolog of Csn5/Jab1 and four uncharacterized PCI components, that interact in a manner suggesting they form a complex analogous to the CSN in S. cerevisiae. These newly identified subunits play a role in adaptation to pheromone signaling. Deletants for individual subunits enhance pheromone response and increase mating efficiency. Overexpression of individual subunits or a human homolog mitigates sst2-induced pheromone sensitivity. Csi1, a novel CSN interactor, exhibits opposite phenotypes. Deletants also accumulate Cdc53/cullin in a Rub1-modified form; however, this role of the CSN appears to be distinct from that in the mating pathway.
Introduction
The Cop9 signalosome (CSN) is an eight-component complex, which is apparently found in all multicellular eukaryotes as a regulator of signaling and developmental processes (Bech-Otschir et al., 2002; Chamovitz and Glickman, 2002). Mutations in CSN subunits cause plant seedlings grown under darkness to mimic light development growth, while Drosophila mutants do not develop beyond larval stage (Chamovitz et al., 1996; Oron et al., 2002). The CSN has been implicated both in the phosphorylation of numerous regulatory proteins, such as Jun and p53 (Seeger et al., 1998; Naumann et al., 1999; Bech-Otschir et al., 2001), and in the removal of Nedd8/Rub1 ubiquitin-like modification from the cullin subunit of E3 ubiquitin ligases (Lyapina et al., 2001; Zhou et al., 2001). Significant structural and genetic similarities are shared between the CSN, lid and eukaryotic translation initiation factor 3 (eIF3) (Glickman et al., 1998; Kapelari et al., 2000; Fu et al., 2001; Glickman and Ciechanover, 2002). All CSN and lid subunits contain one of two signature motifs: the PCI (proteasome, CSN, eIF3) or MPN (Mpr1, Pad1 N-terminal) domains (Aravind and Ponting, 1998; Hofmann and Bucher, 1998; Maytal-Kivity et al., 2002). So far, no evidence for a complex in Saccharomyces cerevisiae has been produced, and straightforward identification of the PCI-motif subunits has been unsuccessful, leading to the assumption that CSN orthologs are not present in this organism (Aravind et al., 2000).
The paradigm of cell-to-cell communication is the mating response in yeast. Haploid cells respond to pheromone by arresting in G1, forming polarized extensions (shmoos), and eventually fusing with their partner to yield a diploid zygote (Elion, 2000; Dohlman, 2002). Pheromone excreted by one mating type cell binds to a G protein-coupled cellsurface receptor of an opposite mating type. The resulting conformational change causes Gα to bind GTP and release Gβγ, which in turn activates the mitogen-activated protein kinase (MAPK) signaling pathway and the synthesis of proteins necessary for the mating process. Desensitization is a vital property of any signaling pathway. Cells that respond to pheromone too easily will arrest prematurely and will be out-competed by cells that do go on dividing as usual. Sst2 is a potent regulator of G-protein signaling (RGS) that desensitizes the mating signal by activating the GTPase activity of free Gα, allowing it to rebind Gβγ (Elion, 2000; Dohlman, 2002). We describe proteins resembling CSN subunits in S. cerevisiae that also participate in the mating pathway by desensitizing the signal and augmenting adaptation to pheromone exposure.
Results and Discussion
Identification of a CSN in budding yeast
In an attempt to determine whether functional homologs of CSN subunits exist in S. cerevisiae, we used the 'generalized profile method' (Hofmann, 2000) to scan the genome for orphaned PCI proteins (see Supplementary data available at EMBO reports Online). When tested for pairwise interactions, three previously uncategorized PCI proteins (Csn9, Csn10 and Csn12) interact with each other and with a protein homologous to Csn5 (Figure 1A). Importantly, plant and yeast Csn5 maintain identical interaction patterns with other yeast CSN candidates. Furthermore, Csn5, Csn9, Csn10 and Pci8/Csn11 coprecipitate together, indicating that they are complexed (Gavin et al., 2002). For comparison, an additional PCI protein, Thp1, is involved in transcription elongation but has not been assigned to any known PCI complex (Gallardo and Aguilera, 2001) and does not interact with the newly identified CSN subunits (Figure 1A). Other PCI-containing proteins such as lid components Rpn3, Rpn5, Rpn6, Rpn7 and Rpn12 or the eIF3 subunits Rpg1/eIF3a and Nip1/eIF3c are essential and classified as components of known PCI complexes. We included in our screen a protein termed Csi1 (CSN interactor 1) that contains neither a PCI nor an MPN domain but can coprecipitate with members of this complex (Uetz et al., 2000; Gavin et al., 2002). Csi1 is found to interact with Csn9 and Csn12 (Figure 1A).
Figure 1.
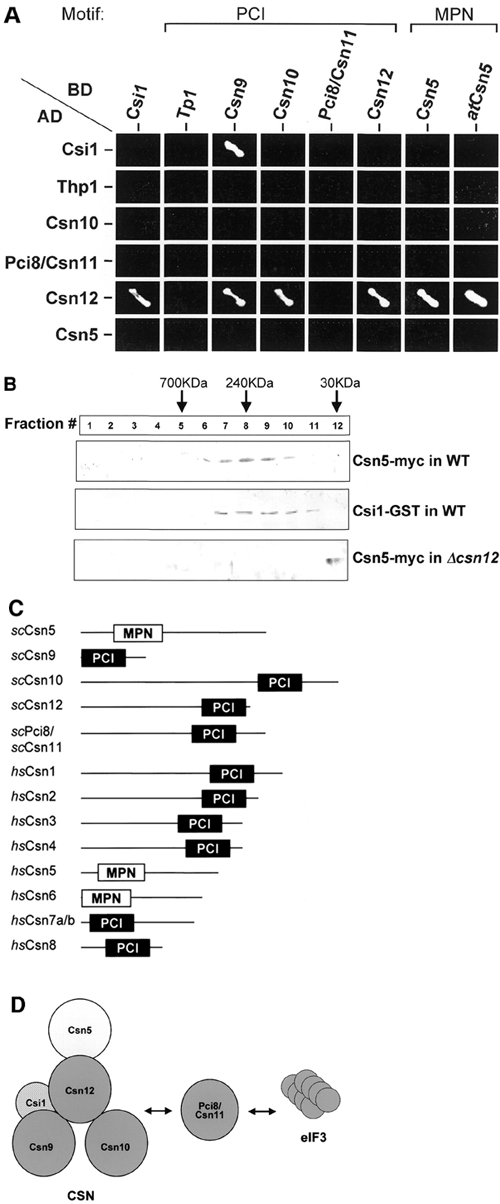
Identification of CSN-like components in S. cerevisiae. (A) Y2H. Pairwise interactions of budding yeast PCI proteins (Csn9, Csn10, Csn12, Pci8/Csn11, Thp1), an MPN protein (Csn5/Rri1) and Csi1. Growth on -ade-his selective media indicates positive interactions. Positive interactions are depicted in (D). Csn5 from plant, atCsn5, is included for comparison. (B) Comigration. Glycerol gradient fractionation of yeast cell extract containing naturally abundant tagged Csn5 or Csi1 in wild-type (WT) or Δcsn12 backgrounds. Csn5 and Csi1 comigrate in a complexed form with an apparent molecular weight of 240 kDa. In Δcsn12, Csn5-myc is detected only in the lowest molecular weight fraction. (C) Structural comparison of identified CSN subunits from budding yeast and human. Cartoons are drawn to scale and depict the signature PCI and MPN domains. See also Supplementary data. (D) Possible model for a CSN-like complex in budding yeast. Csn5, Csn9, Csn10 and Csn12 co-interact with Csn12 playing a pivotal role in the structural organization of this complex (PCI, dark gray; MPN, light gray). Only interactions identified in (A) and (B) are specified. An additional PCI protein, Pci8/Csn11, interacts both with the CSN and eIF3 and may be a shared subunit. A non-PCI/non-MPN protein, Csi1, interacts with Csn9 and Csn12.
Endogenously expressed Csn5-6myc migrates in a complexed form with an apparent molecular weight of ∼240 kDa (Figure 1B). Upon deletion of csn12, Csn5 migrates consistent with its monomeric molecular weight, indicating that incorporation of Csn5 into the complex is dependent on Csn12. Endogenous Csn12 fractionates at ∼240 kDa as well (data not shown). This role for Csn12 in maintaining the integrity of the CSN complex is supported by its y2h interactions (Figure 1A). Similarly in plants, atCsn5 is also incorporated into the CSN only in the presence of atCsn1 (Kwok et al., 1998). As Csi1 does not contain the signature PCI or MPN domains, we tested whether it too fractionates with the CSN. Csi1–GST comigrates with Csn5 in similar molecular weight fractions (Figure 1B).
Four PCI proteins (Csn9, Csn10, Csn11 and Csn12), one MPN protein (Csn5) and one interactor (Csi1) co-interact in S. cerevisiae, whereas the CSN in other eukaryotes forms a 6+2 complex (Figure 1C and D). Despite low sequence homology, dendrogram analysis points to a common evolutionary ancestor of all CSN subunits from yeast and other species (see Supplementary data). Pci8/Csn11 has also been identified as an interactor of the eIF3 complex and a possible homolog of Int6/eIF3e (Shalev et al., 2001), suggesting that it may be a shared subunit. Association of eIF3e with the CSN also occurs in plants and mammals (Yahalom et al., 2001; Hoareau et al., 2002), though the mechanistic meaning of this dual interaction is still unclear. The combined molecular weight of these 'core' subunits is calculated to be ∼250 kDa, in agreement with their migration pattern, suggesting that they form a complex. We cannot preclude possible dimerization of this putative complex, especially as Csn12 interacts with itself, or association of yet additional subunits or interactors (CSIs).
Properties of the CSN
Cellular localization of endogenously expressed GFP-tagged Csn5 is predominantly nuclear (Figure 2A). Nuclear localization is largely maintained upon overexpression (Figure 2B). Deletion of csn12 causes dramatic delocalization of Csn5 throughout the cytoplasm in either case (Figure 2A and B). Nuclear localization has been documented for Csn5 in other species, while compromising complex integrity promotes delocalization of Csn5 in plants (Kwok et al., 1998; Seeger et al., 1998; Mundt et al., 2002). Together, we advocate a nuclear complex with possible cytoplasmic localization for certain monomers or subcomplexes.
Figure 2.
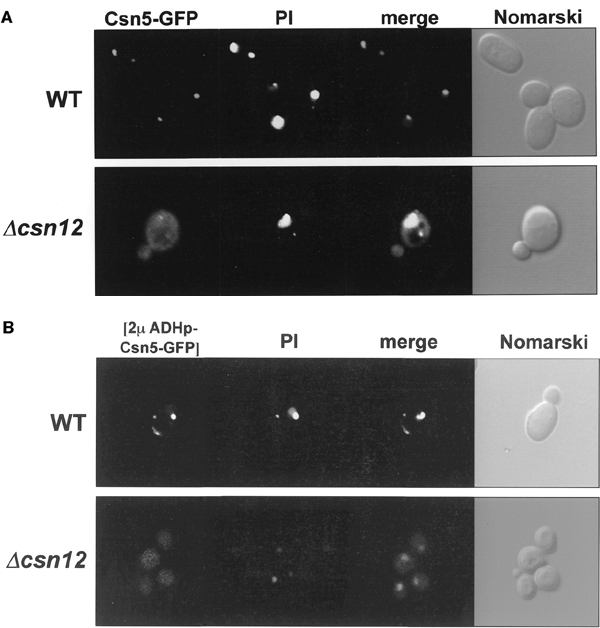
Cellular localization of Csn5 in S. cerevisiae. (A) Localization of natural abundance Csn5. Logarithmically growing cells expressing naturally abundant GFP-tagged Csn5 were monitored by confocal microscopy. Images show Csn5–GFP fluorescence, propidium iodide-stained nuclei (PI), overlay of the two channels, and a Nomarski optical image. Csn5 is concentrated in the nucleus in wild-type (WT) but is delocalized throughout the cell in absence of Csn12. (B) Localization of overexpressed Csn5. Images are as in (A), but strains were transformed with multicopy plasmids expressing GFP-tagged Csn5 under the ADH promotor. Overexpressed Csn5 is similarly localized to the nucleus with slight cellular localization; in Δcsn12, Csn5 is delocalized throughout the cell.
Deletions of csn5, csn9, csn10, pci8/csn11 or csi1 bring about accumulation of Rub1-modified Cdc53/cullin, confirming that the CSN partakes in cullin modification in budding yeast (Figure 3A). While this paper was under review, these results were independently confirmed (Wee et al., 2002), and evidence that Csn5/Rri1 is the catalytic subunit was demonstrated (Cope et al., 2002). Interestingly, Cdc53 modification is insignificantly altered in Δcsn12, which promotes complex disassociation, indicating that integrity of the entire complex is not essential for this hydrolase activity (Figure 3A). Upon overexpression of any individual subunit, however, the extent of Rub1 conjugation is indistinguishable from wild-type (WT; Figure 3B), suggesting that no subunit is independently responsible for Rub1-hydrolase activity. That Csn2 is also essential for cullin deNeddylation in mammalian cells supports a requirement for multiple CSN subunits in deNeddylation (Yang et al., 2002). Quite possibly, removal of Rub1 from Cdc53 may be carried out by (Csn5-containing) subcomplexes or a tightly associated enzyme.
Figure 3.
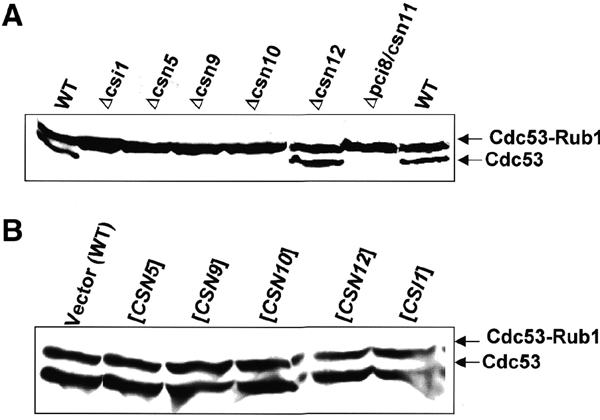
The role of CSN in Cdc53 modification. (A) CSN deletants. Total protein extracts of wild-type (WT) or csn deletants were probed with anti-Cdc53 to visualize the extent of Cdc53 modification by Rub1. Cdc53 accumulates almost exclusively in a Rub1-modified form upon deletion of newly identified CSN-like subunits, with the exception of Δcsn12. (B) Overexpression. Modification of Cdc53 by Rub1 was probed in extracts from cells overexpressing each of the CSN subunits. Cdc53–Rub1 conjugation levels upon overexpression of any CSN subunit are indistinguishable from WT.
The CSN is involved in regulating pheromone response
Befitting a developmental role, deletions of csn5, csn9 or csn12 show increased mating efficiency by up to 100% compared with WT (Figure 4A). Not all subunits behave similarly; deletions of csn10 or pci8 show little or no effect on mating, and Δcsi1 is appreciably poorer at mating than WT (Figure 4A). This effect on mating is linked to greater pheromone sensitivity; most CSN deletants form shmoos quicker and to a larger extent than WT (Figure 4B). Again, Δcsi1 displays an opposite phenotype, distinguishing it from the MPN or PCI members of this complex. Unique phenotypes for individual CSN subunits are documented in other organisms as well (Mundt et al., 2002; Oron et al., 2002).
Figure 4.
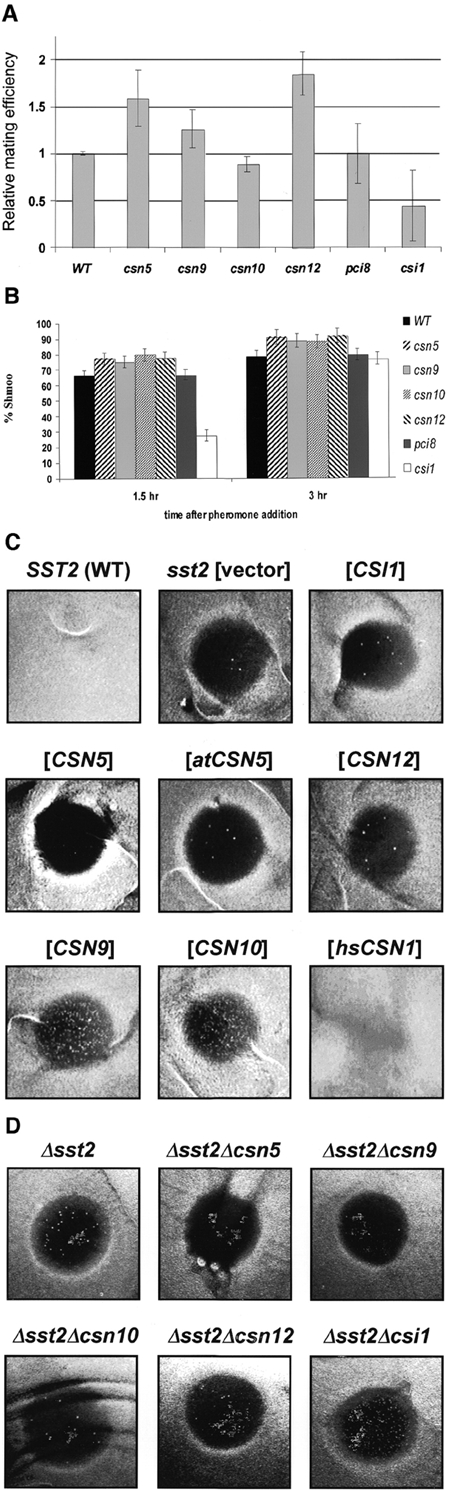
Role of CSN subunits in the mating pheromone pathway. (A) Mating efficiency of CSN disruptions. Deletants in csn5 or csn12 show increased mating efficiency by up to 100% when mated with wild-type (WT) cells. Δcsn10 or Δpci8 show little or no effect on mating, while csi1 null is appreciably poorer at mating compared with WT. Error bars are calculated from an average of 5–7 independent mating experiments for each strain. (B) Shmoo formation. Exponentially growing MATa cultures were treated with 1 mM α-factor pheromone. After 1.5 and 3 h, aliquots were fixed and the percentage of cells with shmoo-like projections were counted. Error bars represent the average of triplicate 200-cell samples. Cells deleted for csn5, csn9, csn10 or csn12 respond to pheromone quicker and to a greater extent than WT cells. Deletion of csi1 delays the response to pheromone exposure. (C) Adaptation to pheromone response (halo assay). Cultures of Δsst2 cells containing a vector plasmid (control) or overexpressing individual CSN subunits were plated on selective media and treated with α-factor to the center of the plate. A halo of no-growth encircles the pheromone due to the inability of sst2 cells to exit G1 arrest. Formation of spontaneous colonies within this halo in Δsst2 culture is at single digits (vector). Overexpression of Csn9, Csn10 or hsCsn1 enhances adaptation, allowing for numerous cells to resume growth within the halo and form colonies. (D) Adaptation to pheromone CSN deletants. As in (C) except that Δsst2 and double deletant strains were plated on complete media. Deletion of csi1 facilitates adaptation to pheromone exposure. Thick growth of Δsst2Δcsi1 occurs at the periphery of the halo, where local concentration of pheromone is lower, and numerous colonies are noticeable throughout the halo. Deletions of other CSN subunits have a mild negative effect on adaptation, decreasing the number of viable colonies compared with the Δsst2 single deletion.
Unable to recover from pheromone-induced cell cycle arrest, Δsst2 cells form a halo of no-growth around the source of pheromone (Figure 4C). Overexpression of Csn9 or Csn10 in Δsst2 allow for numerous cells to adapt and resume growth within this halo (Figure 4C). A strong effect is also observed upon expression of hsCsn1, underscoring a conserved role for PCI members of the CSN. Conversely, deletion of csi1 facilitates adaptation of Δsst2 cells, whereas deletions of CSN subunits (especially Csn5 or Csn9) enhance sensitivity (Figure 4D). It appears that CSN components promote adaptation to pheromone by mitigating G protein-coupled signaling. Interestingly, Csi1 appears to reverse the effect of the CSN on the mating pathway. If so, Csi1 is the first recognized regulator of the CSN in any organism. A link between the CSN and G protein signaling is supported by interaction of Csn9 with Gpa1 (Uetz et al., 2000) and suppression of gpa1 mutations by hsCsn1 (Spain et al., 1996).
It is unclear whether the role of the CSN in the mating response stems from its documented properties. Deletions of most CSN subunits enhance pheromone response and increase Rub1–Cdc53 conjugation levels. However, csn12 null also enhances mating efficiency yet shows no effect on Cdc53 modification, whereas Δcsi1 exhibits an opposite effect on the mating pathway yet accumulates Rub1-modified Cdc53. Notably, overexpressions of individual PCI subunits do not appear to significantly influence levels of Cdc53 modification, yet they decrease pheromone sensitivity, providing evidence for a role of individual subunits in regulating mating but not in hydrolytic removal of Rub1. Together, these results suggest that the role of the CSN in the pheromone response of S. cerevisiae is uncoupled from its role in Cdc53 modification.
G protein-coupled cell surface receptors mediate response to light, small chemicals and hormones. Sst2RGS is considered to be the predominant factor for desensitizing the Gpa1Gα signal in yeast, yet we show that individual CSN subunits can override sst2 deletion. Thus, the CSN negatively regulates this pathway either by aiding in Gα recycling or by repressing downstream components of the MAPK cascade. The conserved function of hsCsn1 in the pheromone response of yeast (Spain et al., 1996), together with the role of CSN in AP-1 signaling (Naumann et al., 1999; Tsuge et al., 2001), suggests that some of the developmental roles of the CSN may also be executed via repression of MAPK pathways.
Methods
Bioinformatics.
Iterative database searches were performed with a non-redundant data set using profiles calculated from previously published alignments (Hofmann and Bucher, 1998) for detection of additional MPN and PCI domains in the S. cerevisiae genome. See Supplementary data for details and results.
CSN subunits.
Rri1 is encoded by YDL216c and is referred to as Csn5 according to the accepted nomenclature (Deng et al., 2000). Due to the lack of a discernible ortholog relationship with characterized CSN subunits 1–8 from other species (see Supplementary data), the suffixes 9, 10, 11 and 12 were used for the putative CSN-like subunits encoded by YDR179c, YOL117w, PCI8/YIL071c and YJR084w, respectively. Csi1 is encoded by YMR025w.
Strains and plasmids.
WT and deletant strains were purchased from EUROSCARF. Double mutants were made by mating haploids, sporulation and tetrad dissection. Relevant genes were cloned by PCR and inserted into CEN plasmids (YCplac111). For overexpression under the ADH2 promotor, open reading frames were cloned into YIplac128. GFP-, GST- or Myc-tagged versions of CSN subunits were constructed in the same plasmids. Based on fluorescence intensity of GFP-tagged proteins, overexpression is ∼10-fold. Eliezer Lifshits and John Colicelli generously provided plasmids with plant CSN5 and hsCSN1, respectively
Yeast 2-hybrid.
Coding regions were inserted into the Matchmaker III yeast 2-hybrid plasmids pGADT7 or pGBKT7 (Clontech). Positive interactions were tested in strain AH109 (Clontech) by Ade and His autotrophy. None of the constructs shown in Figure 1A permitted growth on -ade-his media when transformed alone.
Glycerol gradients.
Total yeast cell extract (0.5 ml) expressing tagged CSN subunits was separated on a 15–40% glycerol gradient in 100 mM NaCl, 50 mM Tris–HCl pH 7.4 and protease inhibitor cocktail (Roche) for 20 h at 30 000 r.p.m. with an SW41 rotor. Gradient fractions (1 ml) were resolved by SDS–PAGE and immunoblotted. For anti-Cdc53 immunoblots (yc-17; Santa Cruz), extract was prepared by mechanical lysis in 50 mM trichloroacetic acid after rinsing with NaF, 10 mM Tris buffer pH 7.4, 0.02% Na-azide and protease inhibitors.
Fluorescence microscopy.
Cells were washed with 70% v/v ethanol and incubated with 0.4 mg/ml RNase A in 50 mM sodium citrate pH 7 for 2 h and transferred to 50 mM sodium citrate pH 7, 10 mM NaCl, 0.1% NP-40 and 5 μg/ml propidium iodide (PI). Reconstructions of GFP and PI channels were made using an MRC-1024 laser confocal scanning microscope (Bio-Rad) with the objective Nikon Plan Apo 603/1.40.
Shmoo formation.
Exponentially growing cells (OD600 = 0.5) were treated with 1 μM final concentration of α-factor pheromone (Sigma). Aliquots were fixed in 7.4% formaldehyde and 0.15 M NaCl. Triplicate batches of 200 cells each were counted by optical microscope for percentage of shmoos.
Mating assay.
Logarithmic cultures of mutant and WT of opposite mating type were incubated at a ratio of 1:10 for 3 h at 30°C and plated on selective media to determine the number of diploid cells. Assays were carried out such that at least 200 diploid cells would be counted in each experiment. The number of haploid cells in each test strain was determined by growth on selective media for the limiting strain.
Halo assays.
Three hundred microliters of exponentially growing culture (OD600 = 0.5) of Δsst2 MATa strains were plated on selective media (for CSN overexpressions) or complete minimal media (for csn deletions) supplemented with 0.1 M citrate, pH 4.5, and 5 μl of 50 μM α-factor was added to the center of the plate. Growth was photographed after 3 days at 30°C.
Supplementary Material
Supplementary data
Acknowledgments
We thank Daniel Kornitzer, Noa Reis, Stavit Drori, Ira Kolotev and Monika Bajorek for suggestions and technical assistance. Suzan Wee and Dieter Wolf are thanked for sharing results and coordinating nomenclature prior to publication. This work was supported by the German Israel foundation for scientific research (GIF), the Israel Science Foundation (ISF), a grant for promotion of research at the Technion and the Wolfson foundation for ubiquitin research.
References
- Aravind L. and Ponting C.P. (1998) Homologues of 26S proteasome subunits are regulators of transcription and translation. Prot. Sci., 7, 1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Watanabe H., Lipman D.J. and Koonin E.V. (2000) Lineagespecific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA., 97, 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D., Kraft R., Huang X., Henklein P., Kapelari B., Pollmann C. and Dubiel W. (2001) COP9 signalosomespecific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J., 20, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D., Seeger M. and Dubiel W. (2002) The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J. Cell Sci., 115, 467–473. [DOI] [PubMed] [Google Scholar]
- Chamovitz D.A. and Glickman M.H. (2002) The COP9 signalosome. Curr. Biol., 12, R232. [DOI] [PubMed] [Google Scholar]
- Chamovitz D.A., Wei N., Osterlund M.T., von Arnim A.G., Staub J.M., Matsui M. and Deng X.W. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant development switch. Cell, 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Cope G.A., Suh G.S., Aravind L., Schwarz S.E., Zipursky S.L., Koonin E.V. and Deshaies R.J. (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science, 298, 608–611. [DOI] [PubMed] [Google Scholar]
- Deng X.W. (2000) Unified nomenclature for the COP9 signalosome: an essential regulator of development. Trends Genet., 16, 202–203. [DOI] [PubMed] [Google Scholar]
- Dohlman H.G. (2002) G proteins and pheromone signaling. Annu. Rev. Physiol., 64, 129–152. [DOI] [PubMed] [Google Scholar]
- Elion E.A. (2000) Pheromone response, mating and cell biology. Curr. Opin. Microbiol., 3, 573–581. [DOI] [PubMed] [Google Scholar]
- Fu H.Y., Reis N., Lee Y., Glickman M.H. and Vierstra R. (2001) Subunit interaction maps for the regulatory particle of the 26s proteasome and the cop9 signalosome reveal a conserved core structure. EMBO J., 20, 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M. and Aguilera A. (2001) A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics, 157, 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Glickman M.H. and Ciechanover A. (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev., 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Glickman M.H. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9/signalosome and eIF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Hoareau A.K., Bochard V., Retty S. and Jalinot P. (2002) Association of the mammalian proto-oncoprotein Int-6 with the three protein complexes eIF3, COP9 signalosome and 26S proteasome. FEBS Lett., 527, 15–21. [DOI] [PubMed] [Google Scholar]
- Hofmann K. (2000) Sensitive protein comparisons with profiles and hidden Markov models. Brief. Bioinform., 1, 167–178. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher P. (1998) The PCI domain: a common theme in three multi-protein complexes. Trends Biochem. Sci., 23, 204–205. [DOI] [PubMed] [Google Scholar]
- Kapelari B., Bech-Otschir D., Hegerl R., Schade R., Dumdey R. and Dubiel W. (2000) Electron microscopy and subunit–subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol., 300, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Solano R., Tsuge T., Chamovitz D.A., Ecker J.R., Matsui M. and Deng X.W. (1998) Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell., 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S. et al. (2001) Promotion of NEDD–CUL1 conjugate cleavage by COP9 signalosome. Science, 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K. and Glickman M.H. (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem., 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt K.E., Liu C. and Carr A.M. (2002) Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell, 13, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M., Bech-Otschir D., Huang X., Ferrell K. and Dubiel W. (1999) COP9 signalosome-directed c-Jun activation/stabilization is independent of JNK. J. Biol. Chem., 274, 35297–35300. [DOI] [PubMed] [Google Scholar]
- Oron E., Mannervik M., Rencus S., Hararisteinberg O., Neuman-Silberberg S., Segal D. and Chamovitz D.A. (2002) COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development, 129, 4399–4409. [DOI] [PubMed] [Google Scholar]
- Seeger M., Kraft R., Ferrell K., Bech-Otschir D., Dumdey R., Schade R., Gordon C., Naumann M. and Dubiel W. (1998) A novel protein complex involved in signal transduction possessing similarities to the 26S proteasome subunits. FASEB J., 12, 469–478. [PubMed] [Google Scholar]
- Shalev A. et al. (2001) Saccharomyces cerevisiae protein Pci8p and human protein eIF3e/Int-6 interact with the eIF3 core complex by binding to cognate eIF3b subunits. J. Biol. Chem., 276, 34948–34957. [DOI] [PubMed] [Google Scholar]
- Spain B.H., Bowdish K.S., Pacal A.R., Staub S.F., Koo D., Chang C.Y., Xie W. and Colicelli J. (1996) Two human cDNAs, including a homolog of Arabidopsis FUS6 (COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammalian cells. Mol. Cell. Biol., 16, 6698–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T., Matsui M. and Wei N. (2001) The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol., 305, 1–9. [DOI] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wee S., Hetfeld B., Dubiel W. and Wolf D.A. (2002) Conservation of the COP9/signalosome in budding yeast. BMC Genet., 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom A., Kim T.H., Winter E., Karniol B., von Arnim A.G. and Chamovitz D.A. (2001) Arabidopsis eIF3e/Int6 associates both with eIF3c and the COP9 signalosome subunit CSN7. J. Biol. Chem., 276, 334–340. [DOI] [PubMed] [Google Scholar]
- Yang X., Menon S., Lykke-Andersen K., Tsuge T., Di Xiao, Wang X., Rodriguezsuarez R.J., Zhang H. and Wei N. (2002) The COP9 signalosome inhibits p27(kip1) degradation and impedes G1–S phase progression via deneddylation of SCF cul1. Curr. Biol., 12, 667–672. [DOI] [PubMed] [Google Scholar]
- Zhou C., Seibert V., Geyer R., Rhee E., Lyapina S., Cope G., Deshaies R.J. and Wolf D.A. (2001) The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem., 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


