Abstract
RNA interference is an evolutionarily conserved double-stranded RNA-triggered mechanism for suppressing gene expression. Rotaviruses, the leading cause of severe diarrhea in young children, are formed by three concentric layers of protein, from which the spike protein VP4 projects. Here, we show that a small interfering RNA corresponding to the VP4 gene efficiently inhibits the synthesis of this protein in virus-infected cells. A large proportion of infected cells had no detectable VP4 and the yield of viral progeny was reduced. Most of the virus particles purified from these cells were triple-layered, but lacked VP4, and were poorly infectious. We also show that VP4 might not be required for the last step of virus morphogenesis. The VP4 gene silencing was specific, since the synthesis of VP4 from rotavirus strains that differ in the target sequence was not affected. These findings offer the possibility of carrying out reverse genetics in rotaviruses.
Introduction
Rotaviruses, the leading cause of severe dehydrating diarrhea in infants and young children worldwide, are non-enveloped viruses formed by three concentric layers of protein that enclose a double-stranded RNA (dsRNA) genome. The innermost layer is formed by protein VP2, which surrounds the viral genome, and small amounts of VP1 and VP3. VP6 constitutes the intermediate layer, and the outermost layer is composed by glycoprotein VP7, which forms the smooth surface of the virus. From this surface, spike-like structures formed by VP4 project (Estes, 1996). VP4 has essential functions in the virus life-cycle, including the attachment of the virus particles to cell receptors and the penetration of the virions into the cell (Estes, 1996).
In infected cells, large cytoplasmic inclusions termed viroplasms are formed, and these are thought to be the sites where double-layered particles (DLPs) assemble. The DLPs then mature by budding from the viroplasm structures into the adjacent endoplasmic reticulum (ER). During this process, which is mediated by the interaction of DLPs with the ER transmembrane rotavirus protein NSP4, the particles acquire a transient membrane envelope (Estes, 1996). This lipid envelope, which contains VP4 and VP7 (Poruchynsky and Atkinson, 1991), is later removed by a largely unknown mechanism, to yield the mature triple-layered particles (TLPs). In addition to the role VP4 may play in the budding process, it has been suggested that this protein may be important for the removal of the lipid membrane from the intermediate enveloped particles (Estes, 1996).
In recent years, an evolutionarily conserved phenomenon called RNA interference (RNAi), which responds to dsRNA by sequence-specific silencing of the homologous genes, has been described. This phenomenon has been observed in invertebrates and plants (Sharp, 2001; Zamore, 2001; Hannon, 2002); however, the demonstration of a RNAi-like response in somatic mammalian cells had been hampered by dsRNA-triggered mechanisms, which nonspecifically inhibit gene expression in vertebrates. The recent demonstration that dsRNA fragments of <30 nucleotides, known as small interfering RNAs (siRNAs), do not activate these mechanisms (Caplen et al., 2001; Elbashir et al., 2001) offers a great opportunity to use this pathway of gene silencing to study the function of mammalian genes. Here, we show that a siRNA corresponding to the rotavirus VP4 gene sequence efficiently and specifically inhibits the synthesis of the VP4 protein in monkey kidney cells, impairing the production of viral progeny. Most rotavirus particles purified from cells transfected with the VP4 siRNA were triple-layered, but lacked VP4, and were poorly infectious. We also show that the absence of VP4 does not prevent the intermediate membrane-enveloped viruses to lose the lipid bilayer.
Results
siRNAVP4 inhibits the production of rotaviral progeny
To determine whether RNAi is functional in monkey kidney cells and its potential effect on rotavirus replication, MA104 cells were lipofected with siRNAs corresponding to gene sequences of either rhesus rotavirus RRV VP4 (siRNAVP4) or protein lamin A/C (siRNALamA/C), which was used as an unrelated control. Twenty-four hours after transfection the cells were infected with RRV, and the virus progeny was harvested 12 h post-infection (p.i.) and its titer determined. The virus produced in cells infected in the presence of siRNAVP4 was ∼25% that of mock-transfected cells, and the viral titer obtained in cells lipofected with siRNALamA/C was not affected (Figure 1A). The titer of viral progeny was reduced in siRNAVP4-transfected cells to a level matching the transfection efficiency of the siRNA (see below).
Figure 1.
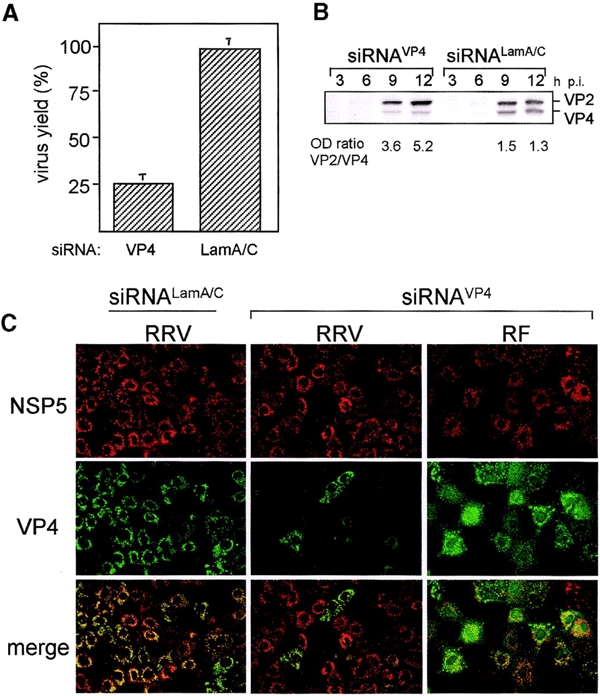
Rotavirus gene silencing. (A) Rotavirus RRV viral progeny produced in cells transfected with siRNAs to VP4 and lamin A/C, expressed as a percentage of the virus yield obtained in mock-transfected cells. The viral progeny titer in mock-transfected cells was 2.1 × 106 ffu/ml. (B) Immunoblot analysis of VP4 synthesized in cells transfected with the indicated siRNA as compared with the synthesis of viral protein VP2, at various times p.i. (C) Immunofluorescence detection of VP4 and the non-structural protein NSP5 in rotavirus RRV- or RF-infected cells in the presence of siRNAVP4 or the unrelated siRNALam A/C.
The synthesis of rotavirus VP4 is affected by siRNAs
Cells lipofected with siRNAs were infected with rotavirus RRV for various times, and the accumulated VP4 protein was detected by immunoblot using a VP4-specific monoclonal antibody (mAb). Viral protein VP2 was used as an internal control. Densitometric analysis of these proteins showed that at 12 h p.i. the VP2/VP4 ratio in cells transfected with siRNALamA/C or siRNAVP4 was 1.3 and 5.2, respectively, indicating that the amount of VP4 that accumulates in the presence of siRNAVP4 is about four times less that of cells treated with the unrelated siRNA (Figure 1B).
To characterize the synthesis of VP4 in individual cells, cell monolayers were lipofected with either VP4 or Lam A/C siRNAs and then infected with rotavirus RRV. Eight hours p.i. the cells were immunostained with a mAb to VP4 and a rabbit polyclonal antibody to NSP5, a non-structural viral protein used as an internal control. In the cells transfected with siRNALamA/C that became infected, both VP4 and NSP5 were readily detected (Figure 1C). In contrast, when the cells were lipofected with siRNAVP4, only a few of the cells stained with the anti-NSP5 antibody were also stained with the VP4 mAb (Figure 1C), indicating that in those cells transfected with the siRNAVP4 the expression of VP4 was reduced to undetectable levels. The VP4-negative cells also expressed regular amounts of two other viral proteins tested, NSP2 and VP7 (data not shown). The fact that some of the cells infected in the presence of siRNAVP4 co-expressed both NSP5 and VP4 very likely reflects the fact that the lipofection efficiency achieved with the siRNAs was ∼75%. This was estimated by determining the percentage of cells with a reduction of the nuclear signal of lamin A/C in cells treated with siRNALamA/C (data not shown). This observation is consistent with the amount of VP4 (∼25% of control) detected by immunoblot in siRNAVP4-treated cells (Figure 1B).
The inhibition of VP4 is specific for rotavirus RRV
To test the sequence specificity of the inhibition of VP4, cells transfected with rotavirus RRV siRNAVP4 were infected with the bovine rotavirus RF or the porcine rotavirus YM, whose VP4 gene sequences differ from that of RRV in five nucleotide positions at the siRNA target site (DDBJ/EMBL/GenBank accession numbers M63231 and U65924 for YM and RF, respectively). The infected cells were then immunostained as described above. In both RF- and YM-infected cells, there was a coincidence in the VP4 and NSP5 signal (shown in Figure 1C for rotavirus RF), showing the sequence specificity of the siRNAVP4 interference. Also, the yield of viral progeny of rotaviruses RF and YM was not reduced in cells treated with RRV siRNAVP4 (data not shown).
Viral particles assembled in the presence of siRNAVP4 lack VP4 and are poorly infectious
To evaluate the effect of reduced levels of VP4 on virus assembly, cells lipofected with siRNAs were infected with rotavirus RRV in the presence of [35S]methionine, and at 24 h p.i. the virus was purified by CsCl density gradients. The virus particles obtained from cells infected in the presence of siRNALamA/C yielded, as regularly observed, two bands (bands 1 and 2 in Figure 2A), which by electrophoresis analysis corresponded to DLPs (formed by proteins VP1, VP2 and VP6) and TLPs (containing, besides the DLP components, proteins VP4 and VP7) (Figure 2B, lanes 1 and 2). In cells infected in the presence of siRNAVP4, these bands (Figure 2A, bands 3 and 5; Figure 2B, lanes 3 and 5) were largely replaced by a new band (Figure 2A, band 4) of intermediate density between DLPs and TLPs. The electrophoretic protein profile of this band showed that the particles with the intermediate density have all proteins present in TLPs, except VP4 (Figure 2B, lane 4). Electron microscopy (EM) analysis of these particles (hereafter called spikeless TLPs) showed that they have a smooth surface, similar to TLPs, and clearly different from DLPs, which have a rough surface appearance (Figure 2C). These findings indicate that VP4 is not needed for the assembly of VP7, although spikeless TLPs consistently showed a larger diameter (∼90 nm) than TLPs (∼80 nm) and DLPs (∼73 nm).
Figure 2.
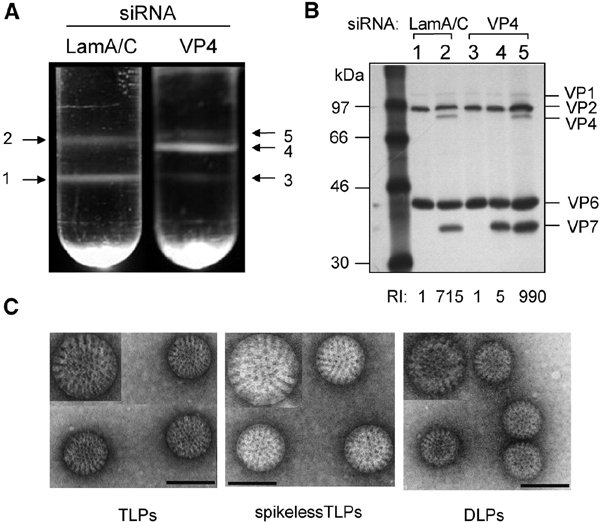
Virus particles synthesized in the presence of siRNAVP4. (A) Isopicnic CsCl gradients of viral particles assembled in the presence of either siRNAVP4 or siRNALamA/C. (B) Gel electrophoresis analysis of the viral particles present in the bands detected in the isopicnic gradients shown in (A). The lane numbers correspond to the number of the band in the gradient. The migration of the viral structural proteins in the gel is indicated. RI refers to the relative infectivity of TLPs and spikeless TLPs as compared with that of DLPs. The infectivities of the particles obtained from siRNALamA/C-treated cells were (ffu/ml) DLPs, 6.7 × 106; TLPs, 4.8 × 109. The infectivities of the particles obtained from siRNAVP4-transfected cells were (ffu/ml) DLPs, 1 × 107; spikeless TLPs, 4.8 × 107; and TLPs 9.9 × 109. (C) EM analysis of DLPs, TLPs and spikeless TLPs purified from siRNAVP4-treated cells, corresponding to bands 3, 4 and 5 of the gradient shown in (A). Magnification ×85 000. The inserts show viral particles amplified 1.5-fold. Scale bars, 90 nm.
Determination of the relative infectivity of the different particles (bottom of panel 2B) showed that, whereas TLPs were ∼1000-fold more infectious than DLPs, the infectivity of spikeless TLPs was similar to that of DLPs. The infectivity of the two latter types of particles is most likely the result of contaminating TLPs, since it was abolished by incubation with neutralizing antibodies to VP4 (data not shown). These results confirm the essential role of VP4 in the infectivity of the virus.
VP4 does not seem to be required for the last steps of virus morphogenesis
The ultrastructural morphology of RRV-infected cells in the presence of either siRNALamA/C or siRNAVP4 was examined by thin EM at 8 h p.i. (Figure 3). In both cases, typical electrodense viroplasm structures were evident in the cytoplasm in close apposition to the ER membrane. DLPs in the periphery of viroplasms were observed to bud into the lumen of the ER, leading to the formation of membrane-enveloped particles, and many of the viral particles observed within the ER had lost their lipid envelope. Given the transfection efficiency of ∼75%, at least 20 siRNAVP4-treated infected cells were analyzed; in all of them, abundant non-enveloped viral particles were observed within the ER. Altogether, these data suggest that VP4 is not required for the budding of DLPs into the ER or for the enveloped intermediate particles to lose their lipid envelope.
Figure 3.
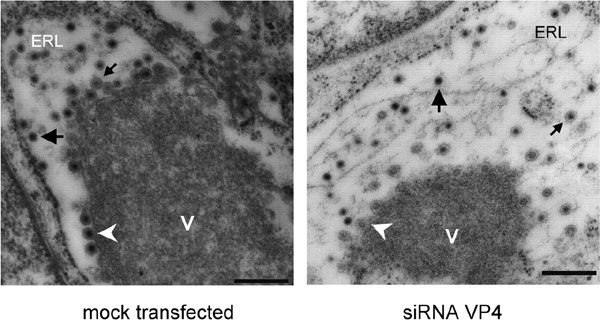
Morphogenesis of RRV rotavirus particles in the presence of mock-transfected or siRNAVP4-transfected MA104 cells. Dense viroplasmic inclusions (V) are abundant in the cytoplasm of rotavirus-infected cells, adjacent to the ER. From these structures DLPs bud (arrowheads) into the lumen of the ER (ERL), resulting in membrane-enveloped particles (small arrows), which later lose the membrane to produce mature triple-layered virions (large arrows). The pictures shown are representative of at least 20 different virus-infected cells. Magnification ×14 000. Scale bars, 400 nm.
Discussion
In this work, we have shown that RNAi efficiently silences the expression of the rotavirus RRV VP4 gene during infection of monkey kidney epithelial cells. This conclusion is based on the fact that, in cells transfected with siRNAVP4, (i) VP4 could not be detected in a large proportion of infected cells (Figure 1C); (ii) the amount of VP4 synthesized was ∼25% that of control cells (Figure 1B); (iii) the viral progeny was reduced by 75% as compared with cells transfected with siRNALamA/C (Figure 1A); and (iv) TLPs lacking VP4 were assembled (Figure 2). The fact that, in the presence of siRNAVP4, VP4 was not detected in a large number of RRV-infected cells, suggests there was a complete, or nearly complete, silencing of the VP4 gene. The silencing of VP4 was not the result of a general antiviral response activated by siRNAs, since the synthesis of other viral proteins was not affected, as detected by immunoblot and immunofluoresence analyses. The specificity of the interference was corroborated by the lack of inhibition of VP4 synthesis in cells infected with rotaviruses RF and YM and by the unaffected replication of these viruses in siRNAVP4-treated cells.
The findings presented in this work have allowed us to evaluate the role of VP4 in the morphogenesis of rotaviruses. In infected cells, the membrane of the ER is modified with viral glycoproteins VP7 and NSP4. VP7 is an integral protein with a luminal orientation, whereas NSP4 is a transmembrane protein, with a cytoplasmic tail that binds the DLPs assembled in the viroplasm, and drives the budding of these particles into the lumen of the ER (Estes, 1996). On the other hand, VP4 is thought to be located in the cytoplasm, possibly between the ER and the viroplasm (Petrie et al., 1982; González et al., 2000). It has been shown that NSP4 and VP7 form a ternary complex with VP4, which has been suggested to participate in the budding process (Maass and Atkinson, 1990). As result of this event, membrane-enveloped particles containing VP4, VP7 and NSP4, in addition to the DLP components, form (Poruchynsky and Atkinson, 1991). As the last step in the morphogenesis of the virions, the enveloped particles in the lumen of the ER lose the membrane envelope by a mechanism that is largely unknown, yielding the mature triple-layered virions. It has been suggested that a hydrophobic domain present in VP4, which is able to interact with, and permeabilize, membranes (Dowling et al., 2000) might have a role during the removal of the lipid envelope (Estes, 1996).
Our results suggest that VP4 is not required for the budding process or for the removal of the lipid membrane from the enveloped particles, since both enveloped and non-enveloped viruses were observed by EM inside the lumen of the ER in cells treated with siRNAVP4, and non-enveloped particles containing VP7, but not VP4, were purified from these cells. Finally, the fact that spikeless TLPs were formed efficiently clearly show that the assembly of VP7 on DLPs is a process independent of the assembly of VP4. However, the larger diameter of the spikeless TLPs as compared with TLPs suggests that, in the absence of VP4, VP7 assembles in a loose manner, probably different from its organization on infectious virions, or that VP4 is needed to tighten the structure of the viral particle.
The highly specific and efficient silencing of rotavirus gene expression by RNAi is remarkable given the highly lytic and rapid replication cycle of the virus, which is completed in ∼12 h, and it opens up the possibility of carrying out reverse genetics in mammalian dsRNA viruses to study gene function, a task that has been elusive for a long time in the field. The specific antiviral effect of siRNA was first demonstrated by Bitko and Barik (2001) in respiratory syncytial virus. Subsequently, such effects were reported in poliovirus (Gitlin et al., 2002) and human immunodeficiency virus (Lee et al., 2002; Novina et al., 2002). Thus, the inhibition of viral gene expression by siRNAs offers the potential for a novel therapeutic approach for rotavirus and other virus infections, once efficient methods for in vivo delivery of siRNAs are developed.
Methods
Cells, viruses and antibodies.
The rhesus monkey epithelial cell line MA104 was grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and was used for all experiments carried out in this work. Rhesus rotavirus RRV was obtained from H.B. Greenberg (Stanford University, Stanford, CA); bovine rotavirus RF was kindly provided by J. Cohen (INRA, Jouy-en-Josas, France); porcine rotavirus YM was isolated in our laboratory. All rotavirus strains were propagated in MA104 cells as described previously (Pando et al., 2002). mAbs to VP2 (mAb 3A8) and VP4 (mAb HS2) were kindly provided by H.B. Greenberg. Rabbit polyclonal serum to NSP5 has been described previously (González et al., 2000).
siRNAs sequence and transfection.
siRNAVP4 had the sequence ucuagguccuuuugcucaatt (sense), uugagcaaaaggaccuagatt (antisense), corresponding to nucleotides 117–135 of the RRV VP4 gene (DDBJ/EMBL/GenBank accession number AY033150). The sequence of siRNALamA/C has been reported previously (Elbashir et al., 2001). The siRNAs were obtained from Dharmacon Research (Lafayette, CO), as annealed duplexes. Transfection of siRNAs was carried out in nearly confluent cell monolayers using 3 μl of oligofectamine (Invitrogen) per 100 μl of siRNAs at 600 pmol/ml in MEM. The transfection mixture was added to cells previously washed with MEM and incubated for 2–3 h at 37°C. After this time the transfection mixture was removed, and the cells were washed with MEM and kept in this medium for 72 h at 37°C before virus infection, except where a different time is indicated.
Infection of cells and titration of viral progeny.
Cell monolayers, in 12- or 24-well plates, were infected with 0.5 virus focus forming units (ffu) per cell, as described previously (Pando et al., 2002), and then incubated for 12 h at 37°C. At this time the cells were then lysed by two freeze–thaw cycles, and the lysates were treated with 10 μg/ml of trypsin for 30 min at 37°C. The infectious titer of the viral preparations was obtained by an immunoperoxidase focus assay (Pando et al., 2002).
Immunoblots.
Cells were transfected with siRNAs and infected with rotavirus RRV as described above. At the indicated times p.i. the cells were lysed, the proteins separated by SDS–PAGE and transferred to nitrocellulose. The transferred proteins were incubated with a mixture of mAbs 3A8 to VP2 and HS2 to VP4, and the bound antibodies were developed by incubation with a peroxidase-labeled anti-mouse immunoglobulin (Ig) (Zymed) and the Western Lightning system (Perkin-Elmer).
Immunofluorescence.
Eight hours p.i. the cells were fixed with 2% paraformaldehyde and permeabilized with 0.5% Triton X-100. The cells were then incubated with a mixture of mAb HS2 and rabbit polyclonal serum to NSP5, followed by staining with goat anti-mouse IgG coupled to Alexa 488 and goat anti-rabbit IgG coupled to Alexa 568 (Molecular Probes). Lamin A/C was detected with mAb 636 (Santa Cruz Biotechnology). The slides were analyzed with a Bio-Rad MRC-600 confocal microscope and CoMOS MPL-1000 software.
Radiolabeling, isolation and analysis of viral particles.
Cells grown in 75 cm2 flasks were transfected with the indicated siRNA and then infected with 1 ffu of rotavirus RRV per cell; viral proteins were labeled for 18 h with 50 μCi/ml [35S]methionine (Dupont NEN) beginning at 3 h p.i. The viral particles were purified by isopicnic CsCl ultracentrifugation, as described previously (Pando et al., 2002). The opalescent bands corresponding to the different types of particles were collected by punction, and the protein composition of each band analyzed by SDS–PAGE and fluorography. To determine the infectivity of each fraction, CsCl was removed by desalting-centrifugation on Sephadex G-25, the virus was activated with trypsin, and the infectious titer was determined as described above.
Electron microscopy.
Cells transfected with siRNAVP4 or mock-transfected were infected with 0.5 ffu of rotavirus RRV per cell, and 8 h p.i. the cells were detached from the culture plates, fixed in 2.5% glutaraldehyde/0.1 M cacodylate, embedded in LR-white resin and postfixed with 1% osmium tetroxide. Thin sections obtained were stained with 0.5% uranyl acetate/1% lead citrate. To analyze the purified virus particles, the fractions were desalted as described above, and a drop was applied to a carbon-coated copper grid and then negative stained with uranyl acetate. The grids were examined in a Zeiss EM-900 electron microscope at 80 kV.
Acknowledgments
We are grateful to Xóchitl Alvarado and Lorena López for their excellent technical assistance with the confocal and electron microscopes, respectively. This work was partially supported by grants 55000573 and 55000613 from the Howard Hughes Medical Institute, G37621N from Conacyt and 262-M from Conacyt-SSA, Mexico.
References
- Bitko V. and Barik S. (2001) Phenotypic silencing of cytoplasmic genes using sequencespecific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol., 1, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen N.J., Parrish S., Imani F., Fire A. and Morgan R.A. (2001) Specific inhibition of gene expression by small doublestranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling W., Denisova E., LaMonica R. and Mackow E.R. (2000) Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol., 74, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K. and Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Estes M.K. (1996) Rotaviruses and their replication. In Fields, B.N. et al. (eds), Virology. Raven Press, New York, Vol. 2, pp. 1625–1655. [Google Scholar]
- Gitlin L., Karelsky S. and Andino R. (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature, 418, 430–434. [DOI] [PubMed] [Google Scholar]
- González R.A., Espinosa R., Romero P., López S. and Arias C.F. (2000) Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol., 145, 1963–1973. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Lee S.N., Dohjima T., Bauer G., Li H., Li M.-J., Ehsani A., Salvaterra P. and Rossi J. (2002) Expression of small interfering RNAs targeted againts HIV-1 rev transcripts in human cells. Nat. Biotech., 19, 500–505. [DOI] [PubMed] [Google Scholar]
- Maass D.R. and Atkinson P.H. (1990) Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J. Virol., 64, 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina C.D. et al. (2002) siRNA-directed inhibition of HIV-1 infection. Nat. Med., 8, 681–686. [DOI] [PubMed] [Google Scholar]
- Pando V., Isa P., Arias C.F. and López S. (2002) Influence of calcium on the early steps of rotavirus infection. Virology, 295, 190–200. [DOI] [PubMed] [Google Scholar]
- Petrie B.L., Graham D.Y., Hanssen H. and Estes M.K. (1982) Localization of rotavirus antigens in infected cells by ultrastructural immuno-cytochemistry. J. Gen. Virol., 63, 457–467. [DOI] [PubMed] [Google Scholar]
- Poruchynsky M.S. and Atkinson P.H. (1991) Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol., 65, 4720–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference-2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Zamore P. (2001) RNA interference: listening to the sound silence. Nat. Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]


