Abstract
In bacteria, the Min system plays a role in positioning the midcell division site by inhibiting the formation of the earliest precursor of cell division, the Z ring, at the cell poles. However, whether the Min system also contributes to establishing the precise placement of the midcell Z ring is unresolved. We show that the Z ring is positioned at midcell with a high degree of precision in Bacillus subtilis, and this is completely maintained in the absence of the Min system. Min is therefore not required for correct midcell Z ring placement in B. subtilis. Our results strongly support the idea that the primary role of the Min system is to block Z ring formation at the cell poles and that a separate mechanism must exist to ensure cell division occurs precisely at midcell.
Introduction
During vegetative growth, cell division in rod-shaped bacteria like Escherichia coli and Bacillus subtilis occurs at the cell centre. The earliest event in division is the assembly of the tubulin-like protein FtsZ at the midcell site into an annular structure, called a Z ring. This ring subsequently contracts and is accompanied by the invagination (or constriction) of the cell envelope to generate two newborn cells. Thus, the Z ring defines the position of the division site and is an accurate marker for this site.
It is well known that the Min proteins play a role in positioning the division site (Margolin, 2001a; Rothfield et al., 2001). These proteins are not essential for viability, but in their absence E. coli and B. subtilis divide at the cell poles, as well as medially, to generate DNA-less minicells (Adler et al., 1967; Reeve et al., 1973). There are three Min proteins in E. coli and B. subtilis (de Boer et al., 1992; Levin et al., 1992; Varley and Stewart, 1992; Lee and Price, 1993). Two of these, MinC and MinD, form a multimeric membrane-associated complex (MinCD) that directly inhibits Z ring assembly (Bi and Lutkenhaus, 1993; Hu et al., 1999; Pichoff and Lutkenhaus, 2001). The third protein, DivIVA in B. subtilis and MinE in E. coli, is required for topological specificity of MinCD inhibition (de Boer et al., 1989; Cha and Stewart, 1997; Edwards and Errington, 1997). In E. coli, the MinCD inhibitor undergoes a MinE-dependent oscillation from pole to pole so that its concentration, over time, is highest at the poles and lowest at the cell centre (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b; Fu et al., 2001; Hale et al., 2001). In B. subtilis, the MinCD complex does not appear to oscillate. It remains concentrated in the two polar regions through interaction with the pole-anchored DivIVA (Edwards and Errington, 1997; Marston et al., 1998; Marston and Errington, 1999). Hence, it is clear how the Min system can specifically block Z ring formation in the polar regions of cells of both organisms. Presumably, this is needed because of ineffective or reduced nucleoid occlusion (inhibition) of Z ring formation in the nucleoid-free polar regions (Harry, 2001; Margolin, 2001b).
Recently, the possibility has been raised that the Min system in E. coli acts as a centre-finding tool (Hale et al., 2001). The midcell site equidistant between the two poles would be coincident with the lowest concentration of the MinCD inhibitor, which would be recognized to trigger Z ring assembly. Mathematical models are consistent with this idea (Meinhardt and de Boer, 2001; Kruse, 2002). In the absence of the Min system, both E. coli and B. subtilis cells can divide between two replicated nucleoids (Levin et al., 1998; Yu and Margolin, 1999), but no unambiguous quantitative assessment of a role for Min in the precision of midcell Z ring positioning has been made. In fact, it is still not known how precise 'midcell' Z ring positioning is in wild-type B. subtilis cells, so it is not clear whether the Z rings that formed between replicated nucleoids in B. subtilis MinCD− cells (Levin et al., 1998) were positioned less precisely than those in wild-type cells. The crucial unanswered question remains: does the Min system contribute to establishing midcell site positioning?
In the present work, we used immunofluorescence microscopy (IFM) and FtsZ–YFP detection to examine the position of the first Z ring to form in cells growing out from spores of B. subtilis, in the presence or absence of the MinCD inhibitor. The synchronous spore system is ideal because, unlike exponentially growing cells of MinCD− strains, one can ensure that the possible formation and removal of a minicell from one end has not influenced the relative position within the rod of the first midcell Z ring. We show that the midcell Z ring in wild-type B. subtilis is defined with a high degree of precision, and this is completely maintained in the absence of MinCD. Thus, in B. subtilis at least, the Min system is not needed to establish the precise position of cell division within the central region. Apparently, the primary role of the Min system is to prevent division from occurring very close to the cell poles.
Results and discussion
Z ring positioning in the absence of MinCD using IFM
We initially used IFM to measure Z ring position in outgrown spores of B. subtilis. Strain dna-1 (thyA thyB dnaB1) was used as a background MinCD+ strain in these experiments because it has been well characterized using the spore germination system and these spores are very synchronous (Harry et al., 1999; Regamey et al., 2000). Notably, the dnaB1 mutation has no effect on Z ring positioning at the permissive temperature of 34°C (M. Isaacs and E.J. Harry, unpublished data). Min proteins are recruited to the cell poles very soon after germination of B. subtilis dna-1 spores, prior to formation of the first Z ring, indicating that the Min system plays a role in positioning this ring (E.J. Harry and P.J. Lewis, unpublished data). Outgrown spores of dna-1 were collected at 160 min for Z ring visualization using IFM. The results are shown in Figure 1A, where Z ring position is expressed as the distance of the Z ring from the nearest pole divided by cell length. For statistical calculations, all measurements of centrally located Z ring positions were randomized with respect to the pole used to measure this pole–Z ring distance. A Z ring was detected in 71% of cells (181 cells scored) with an average position of 0.50 ± 0.030, with a standard deviation of 6.0% off centre.
Figure 1.
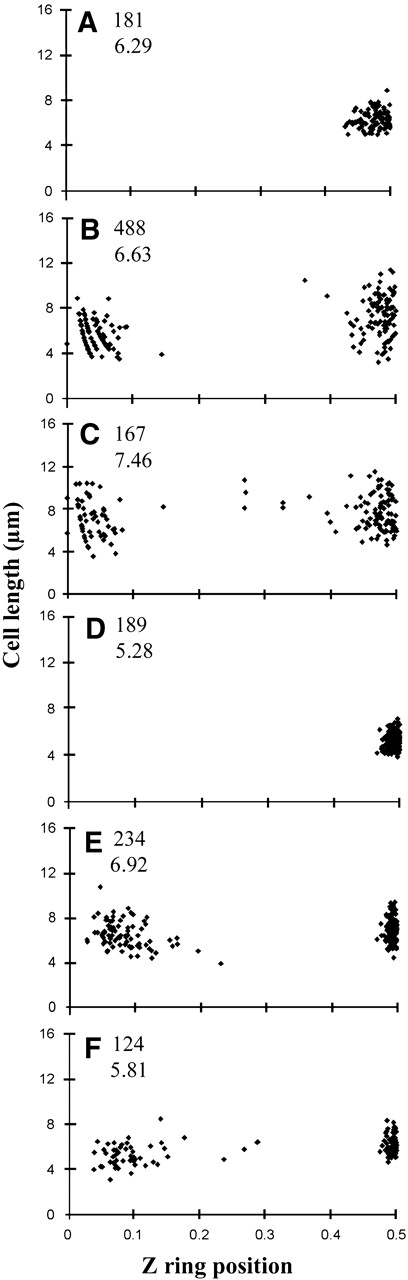
Z ring position in B. subtilis cells grown out from spores or vegetatively growing cells. Z ring position is given by the ratio of the shorter distance from a cell pole to the total cell length. (A) FtsZ distribution in outgrown spores of dna-1 (MinCD+) processed for IFM. (B) FtsZ distribution in the absence of MinCD in outgrown spores of SU429 processed for IFM. (C) FtsZ distribution in the absence of MinCD in vegetative cells of SU429 processed for IFM. (D) FtsZ–YFP distribution in live outgrown spores of strain SU434 (MinCD+). (E and F) FtsZ–YFP distribution in the absence of MinCD in live outgrown spores of SU433 and SU440, respectively. The additional numbers show the number of cells scored (upper) and the mean cell length in micrometres (lower).
The minC and minD genes in the dna-1 strain were inactivated by insertion of specR into the minCD locus, which deletes all of minC and all but the last two codons of minD (Levin et al., 1998), to give strain SU429. IFM of outgrown SU429 cells (collected at 180 min) showed that 54% of them contained Z rings (488 cells scored). (The lower frequency of Z rings is likely due to differences in permeabilization efficiencies after fixation for IFM.) Of cells containing Z rings, 77% had a single ring, 21% had two rings and 1.5% had more than two rings. Z rings were either positioned close to or at the cell centre or near the cell pole (Figure 1B). In cells containing a single Z ring, the frequencies of polar rings and centrally located rings were 49 and 51%, respectively. The decreased proportion of centrally located rings compared with that observed previously (89%) for cells grown in minimal medium (Levin et al., 1998) possibly reflects the shorter time interval between initiation of each round of DNA replication in the richer germination medium, resulting in less DNA-free space and increased nucleoid occlusion in this region of the cell. The proportion of central Z rings did increase to ∼70% in more slowly growing vegetative cells of SU429 (see below). The position of the Z ring was measured only in cells that contained one ring. The average central ring position was 0.50 ± 0.034, with a standard deviation of 6.7% off centre. Thus, Z ring positioning in the central region is just as precise as in MinCD+ outgrown cells (compare Figure 1A with B). These results strongly suggest that the Min system is not required for the precise positioning of a Z ring at midcell. We also examined the precision of central Z ring positioning in SU429 cells (MinCD−) during vegetative growth (minimal medium) and obtained a similar result. In these experiments, we measured the position of Z rings in cells containing one ring (90% of cells) that were of similar length to wild-type cells so that longer cells, which would have more than one potential division site, were omitted from the analysis. The average central ring position was 0.50 ± 0.044, with a standard deviation of 8.9% off centre (Figure 1C).
Midcell Z ring positioning is highly precise
Using IFM, a significantly higher standard deviation for Z ring position in wild-type B. subtilis outgrown cells (6.7% off centre) was obtained than that reported for E. coli (2.6% off centre; Yu and Margolin, 1999). This may be due to the slight swelling of B. subtilis cells that occurs during processing for IFM (Harry et al., 1999). We therefore used live (unfixed, unpermeabilized) cells to see if a more precise definition of Z ring position could be obtained. We visualized Z rings in live cells grown out from germinated spores of B. subtilis that contained an FtsZ–YFP fusion. Bacillus subtilis strain SU434 contains a xylose-inducible ftsZ-yfp fusion inserted into the chromosome at the amyE locus in addition to the wild-type copy of ftsZ. As reported previously (Levin et al., 1999), the wild-type copy was found to be essential for division. Z ring assembly and cell division appear completely normal in strain SU434. Spores of SU434 were grown out in the presence of xylose and collected at 270 min for Z ring visualization (Figure 2A). This collection time simply reflects the longer time taken for spores of this strain to germinate and grow out than those of strain SU429. Due to the relatively strong autofluorescence of the spore coats, which remained attached to many cells, two differently exposed images were obtained and overlaid so that the lower exposure image could be used to determine the position of the cell ends. FtsZ–YFP was detected in almost all cells (>99%) and usually localized as a discrete band in the cell centre. Very few cells (<2%) contained more than one Z ring and were omitted from analysis. The average Z ring position was 0.50 ± 0.011 (189 cells; Figure 1D). This standard deviation (2.2% off centre) is about one-third that calculated using IFM of outgrown spores and is similar to that in E. coli (2.6%; Yu and Margolin, 1999). Thus, FtsZ–YFP detection in live B. subtilis cells yields a significantly more precise localization of Z rings at the cell centre than IFM. Interestingly, this is significantly more precise than that observed for midcell PolC–GFP localization in B. subtilis vegetative cells (Lemon and Grossman, 1998).
Figure 2.
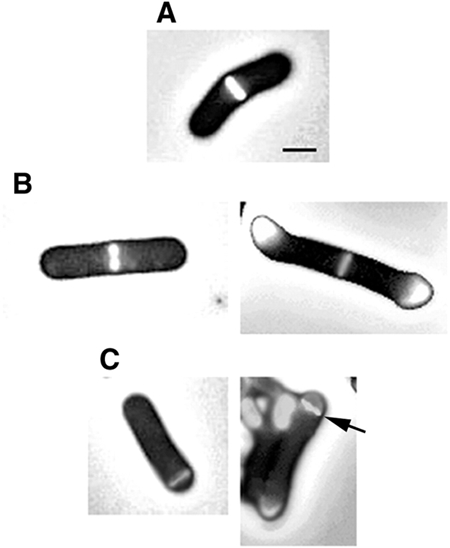
Z rings visualized as an FtsZ–YFP fusion in live cells grown out from B. subtilis spores. (A) A midcell Z ring in SU434 (MinCD+). Midcell Z rings (B) and polar Z rings (C) detected in the MinCD− strain, SU433. The image on the right in (B) and (C) shows autofluorescence of the spore coats attached to the cell ends. The arrow on the right image in (C) shows the polar Z ring. Scale bar, 1 μm.
Z ring positioning in live cells in the absence of MinCD
Having more accurately determined the precision of Z ring positioning at midcell in wild-type (MinCD+) cells, we then examined Z ring positioning in live MinCD− cells. Strain SU433 is genetically identical to SU434, except that it contains the same minCD deletion as in strain SU429. Outgrown spores of SU433 (MinCD−) were collected at 210 min for Z ring visualization. Again, Z rings were detected in almost all cells (>99%), with very few cells containing more than one ring (Figure 2B and C). All rings were positioned either centrally or near the cell pole (Figure 1E). Note the complete absence of Z rings from the ∼0.25–0.47 region of the cell, which probably reflects nucleoid occlusion. The relative frequency of medial and polar rings was 65 and 35%, respectively (234 cells scored). The average medial Z ring position was 0.50 ± 0.008; a standard deviation of 1.6% off centre. Thus, the precision of Z ring positioning in the central region in these MinCD− cells was as least as high as that observed in MinCD+ cells (compare Figure 1D with E). We repeated the above experiments with a completely different deletion of the minC and minD genes (strain SU440), which would remove 107 residues from the C-terminus of MinC and 188 residues from the N-terminus of MinD (Table 1; Lee and Price, 1993). Figure 1F shows the distribution of Z ring positioning in this strain at 210 min. The precision of Z ring positioning in the central region of the cell (0.50 ± 0.009) was the same as for SU433.
Table 1.
B. subtilis strains
| Strain | Genotype | Reference or constructiona |
|---|---|---|
| SB566 | 168 thyA thyB trpC2 | Adler et al. (1967); A.T. Ganesan |
| PL990 | JH642 trpC2 phe ΔminCD990::spc | Levin et al. (1998) |
| PB302 | trpC2 minCD Δ1::cat | Lee and Price (1993) |
| 1058 | 168 trpC2 amyE:: (spc Pxyl-ftsZ-yfp) | Feucht and Lewis (2001) |
| dna-1 | 168 dnaB1(Ts) thyA thyB trpC2 | N. Sueoka |
| SU429 | 168 dnaB1(Ts) thyA thyB trpC2 minCD990::spc | PL990→SU46 (Spec) |
| SU432 | thyA thyB trpC2 ΔminCD990::spc | PL990→SB566 (Spec) |
| SU433 | thyA thyB trpC2 ΔminCD990::spc amyE:: (spc Pxyl-ftsZ-yfp) | 1058→SU432b |
| SU434 | thyA thyB trpC2 amyE:: (spc Pxyl-ftsZ-yfp) | 1058→SB566 (Spec) |
| SU440 | thyA thyB trpC2 minCDΔ1::cat amyE:: (spc Pxyl-ftsZ-yfp) | PB302→SU434 (Cm) |
aFor transformations, the source of the DNA is shown, followed by an arrow, then the recipient strain, with the antibiotic selection, auxotrophic or otherwise in parentheses.
bStrain constructed by congression.
In conclusion, we have shown that positioning of the Z ring within the central region of the B. subtilis cell is highly precise, and this degree of precision is completely maintained in the absence of the MinCD inhibitor. The Min system is therefore not required for the correct placement of a Z ring at midcell in B. subtilis. Our findings strongly support the suggestion that the primary role of the Min system is to prevent Z rings forming close to the poles, where there is reduced or no nucleoid occlusion (Yu and Margolin, 1999; Regamey et al., 2000).
A similar role for the Min system in all bacteria?
Since Min− cells divide either at cell poles or between two segregated chromosomes during vegetative growth, a range of cell sizes is produced, from minicells to cells longer than normal. In a previous study examining Z ring positioning in minCDE-deleted E. coli cells, non-polar Z rings even in short cells were significantly less precisely positioned at the cell centre than Min+ cells of similar length (Yu and Margolin, 1999). However, it is not possible to accurately define midcell even in these short cells, because it is not known what division, polar or midcell, gave rise to these cells. Our findings highlight the advantage of the outgrown spore approach, as it is uncomplicated by previous cell division events. The Min system is not present in all bacteria, nor has it been found to be essential in any organism (Margolin, 2001b). It is therefore quite conceivable that the primary role of the Min system, in all bacteria that have it, is to prevent Z rings forming at the polar regions of the cells and that a separate, conserved mechanism exists to ensure that Z rings are placed precisely at the midcell division site.
How is the midcell site determined?
It is widely acknowledged that the nucleoid negatively regulates bacterial cell division, but the mechanism remains unknown. It has been proposed that, when the nucleoids segregate and move away from the cell centre, the block to division is relieved at this site (Woldringh et al., 1991). More recently, it has been shown that nucleoids can also occlude Z ring formation (Yu and Margolin, 1999). Can relief of nucleoid occlusion be solely responsible for establishing the precision with which a Z ring is placed at midcell in B. subtilis? Studies using outgrown spores of B. subtilis identified a link between progress of the first round of chromosome replication and midcell Z ring assembly (Harry et al., 1999; Regamey et al., 2000). In these studies, DNAstaining experiments under various conditions of DNA replication inhibition suggested that a nucleoid-free space at the cell centre was not required for midcell Z ring formation (Regamey et al., 2000). It was proposed that the midcell anchored replisome is an additional and perhaps crucial factor in defining the precise site of Z ring assembly. However, in the present work, it appears that Z rings are more precisely positioned than the midcell replisome. So it may not be the replisome as such that defines the midcell Z ring assembly site, but, rather, a site or structure to which it is anchored. Further work is needed to clarify the situation.
Methods
Bacterial strains.
Bacterial strains used in this study are listed in Table 1. New strains were constructed using standard techniques.
Growth of strains and spore preparation, germination and outgrowth.
Vegetative B. subtilis SU429 cells were grown at 34°C in S7 medium with 1% (w/v) glucose (Lemon and Grossman, 1998) and harvested at mid-exponential phase (A590 ∼0.5) for IFM. Bacillus subtilis dna-1 spores were prepared as described by McGinness and Wake (1979). All other B. subtilis spores were prepared by the same method, but using fresh potato infusion (0.2 g potato/ml) instead of Difco Potato Extract. Germination (2 × 108 spores/ml) and outgrowth were performed at 34°C in GMD medium (Regamey et al., 2000) supplemented with 20 μg/ml thymine and xylose (0.2% w/v) when required. Cells were harvested when the first Z ring was visible in the majority of cells.
Fluorescence microscopy.
IFM was performed essentially as described previously (Harry et al., 1999), with the following modifications. Glutaraldehyde (0.008%) was added to the fixation mixture and either 7.5 mg/ml (dna-1) or 5 mg/ml (SU429) lysozyme was used. Affinity-purified sheep anti-FtsZ antibodies (diluted 1:400) and affinity-isolated donkey antisheep fluorescein isothiocyanate (FITC)-conjugated antibodies (Jackson ImmunoResearch; diluted 1:100) were used. Live cells were prepared for FtsZ–YFP detection as described in Lemon and Grossman (1998). A U-MWIB (Olympus) excitation cube was used for visualizing fluorescein and FtsZ–YFP.
Acknowledgments
We thank Dr Peter Lewis, Dr Petra Levin and Dr Chester Price for their gifts of B. subtilis strains 1058, PL990 and PB302, respectively. This work was supported by grants from the Australian Research Council.
References
- Adler H.I., Fisher W.D., Cohen A. and Hardigree A.A. (1967) Miniature Escherichia coli cells deficient in DNA. Proc. Natl Acad. Sci. USA, 57, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus J. (1993) Cell division inhibitors, SulA and MinCD, prevent localization of FtsZ. J. Bacteriol., 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J.-H. and Stewart G.C. (1997) The divIVA minicell locus of Bacillus subtilis. J. Bacteriol., 179, 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley R.E. and Rothfield L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley R.E. and Rothfield L.I. (1992) Roles of MinC and MinD in the sitespecific septation block mediated by MinCDE system of Escherichia coli. J. Bacteriol., 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.H. and Errington J. (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol., 24, 905–915. [DOI] [PubMed] [Google Scholar]
- Feucht A. and Lewis P.J. (2001) Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene, 264, 289–297. [DOI] [PubMed] [Google Scholar]
- Fu X., Shih Y.-L., Zhang Y. and Rothfield L.I. (2001) The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl Acad. Sci. USA, 98, 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.A., Meinhardt H. and de Boer P.A. (2001) Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J., 20, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E.J. (2001) Bacterial cell division: regulating Z ring formation. Mol. Microbiol., 40, 795–803. [DOI] [PubMed] [Google Scholar]
- Harry E.J., Rodwell J., and Wake R.G. (1999) Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol., 33, 33–40. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Hu S., Mukherjee A., Pichoff S. and Lutkenhaus J. (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA, 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse K. (2002) A dynamic model for determining the middle of Escherichia coli. Biophys. J., 82, 618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. and Price S. (1993) The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol. Microbiol., 7, 601–610. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman A.D. (1998) Localisation of bacterial DNA polymerase: evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Levin P.A., Margolis P.S., Setlow P., Losick R. and Sun D. (1992) Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol., 174, 6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A., Shim J.J. and Grossman A.D. (1998) Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J. Bacteriol., 180, 6048–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A., Kurster I.G. and Grossman A.D. (1999) Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 96, 9642–9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. (2001a) Bacterial cell division: a moving MinE sweeper boggles the MinD. Curr. Biol., 11, R395–R398. [DOI] [PubMed] [Google Scholar]
- Margolin W. (2001b) Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol., 4, 647–652. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Errington J. (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localisation and activation of MinC. Mol. Microbiol., 33, 84–96. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Thomaides H.B., Edwards D.H., Sharpe M.E. and Errington J. (1998) Polar localisation of the MinD protein of Bacillus subtilis and its role in selection of the midcell division site. Genes Dev., 12, 3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness T. and Wake R.G. (1979) Division septation in the absence of chromosome termination in Bacillus subtilis. J. Mol. Biol., 134, 251–264. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. and de Boer P.A.J. (2001) Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl Acad. Sci. USA, 98, 14202–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S. and Lutkenhaus J. (2001) Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol., 183, 6630–6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer P.A.J. (1999a) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer P.A.J. (1999b) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J.N., Mendelson N.H., Coyne S.I., Hallock L.L. and Cole R.M. (1973) Minicells of Bacillus subtilis. J. Bacteriol., 114, 860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regamey A., Harry E.J. and Wake R.G. (2000) Midcell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: coordinating chromosome replication with cell division. Mol. Microbiol., 38, 423–434. [DOI] [PubMed] [Google Scholar]
- Rothfield L.I., Shih Y.L. and King G. (2001) Polar explorers: membrane proteins that determine division site placement. Cell, 106, 13–16. [DOI] [PubMed] [Google Scholar]
- Varley A.W. and Stewart G.C. (1992) The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J. Bacteriol., 174, 6729–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C.L., Mulder E., Huls P.G. and Vischer N. (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol., 142, 309–320. [DOI] [PubMed] [Google Scholar]
- Yu X.-C., and Margolin W. (1999) FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localisation. Mol. Microbiol., 32, 315–326. [DOI] [PubMed] [Google Scholar]


