Abstract
Fluorescent amplified fragment length polymorphism (AFLP) was applied to 46 Salmonella enterica serovar Typhimurium isolates of Australian origin comprising nine phage types, by using the restriction enzymes MseI and EcoRI and all 16 possible MseI +1-EcoRI +1 primer pair combinations. AFLP in the present study showed a very good discrimination power with a Simpson index of diversity of 0.98, and 35 different AFLP patterns were observed in the 46 isolates. AFLP grouped most serovar Typhimurium isolates by phage type and enabled differentiation of phage types. Furthermore, 84 phage-type-specific polymorphic AFLP fragments, for which presence or absence correlated with phage type (including 25 with one exception to phage type specificity) were observed in the 46 strains studied. Eighteen phage-type-specific AFLP fragments were cloned and sequenced. Fifteen are of known genes or have a homologue in the databases. Three sequences are plasmid related, eight are phage related, and four relate to chromosomal genes. Twelve of the 18 fragments are polymorphic because the DNA is present or absent as indicated by Southern hybridization, and we see good potential to use sequences of these fragments as the basis for multiplex PCR and development of a microarray-based molecular phage-typing method for serovar Typhimurium.
Salmonella enterica has been divided into over 2,000 serovars based on the combination of antigenic properties of flagellar H1 and H2 antigens and the polysaccharide O antigen (29). S. enterica serovar Typhimurium is a very diverse serovar, which includes by definition all 1,4,[5],12:i:1,2 isolates. Serovar Typhimurium causes salmonellosis (Salmonella gastroenteritis) among humans and domestic animals worldwide. It was for many years the serovar commonly isolated, but the other major serovar, Enteritidis, has outnumbered serovar Typhimurium in many areas in recent years (7).
Effective epidemiological surveillance and control of serovar Typhimurium requires the accurate subtyping of strains to determine potential sources of infection. A number of different phenotypic and genotypic methods have been used for this purpose, including phage typing, biotyping, plasmid profile typing, and plasmid fingerprinting. Phage typing is a commonly used method that has proved to be very useful in epidemiological surveillance of serovar Typhimurium infections. The phage-typing scheme is based on combinations of resistance or degree of sensitivity of serovar Typhimurium isolates to a series of specific bacteriophages (phages). The Anderson phage-typing scheme being used today distinguishes 207 definitive phage types (DTs) with 34 phages (3). The use of phage typing has enabled the rise and fall of different forms and geographical distribution to be monitored, with DT104, for example, rising in recent years to dominance in serovar Typhimurium in much of Europe but remaining rare in Australia.
However, knowledge of relationships between phage types is very limited, and the genetic basis of phage type variation remains largely unknown. In some cases, one serovar Typhimurium phage type may be converted to a different type via plasmid, transposon, or temperate phage acquisition (2, 41). Phage type conversion in S. enterica serovar Enteritidis caused by the introduction of a resistance plasmid has also been reported elsewhere (8).
Although phage typing plays an important role in epidemiology, it has some drawbacks. First, a proportion of serovar Typhimurium strains cannot be classified by the present phage-typing scheme. Second, the recording of results is, to a significant extent, subjective, so that misclassification can occur (5).
Molecular typing methods, such as IS200 typing, ribotyping, and pulsed-field gel electrophoresis (PFGE), have also been used to type serovar Typhimurium and/or to define the relationships between and within phage types of serovar Typhimurium (16, 27, 28). However, IS200 typing and ribotyping detect only variations in part of the genome. PFGE detects variations in the whole genome without requiring knowledge of sequence and has been quite widely used, with reports of its use for both primary discrimination of S. enterica isolates and subdivision of phage types.
In recent years, a novel DNA fingerprinting method, amplified fragment length polymorphism (AFLP), has proved itself as a high-resolution genotyping method and a useful tool in taxonomy and epidemiological studies of microorganisms (1, 14, 19). AFLP is based on the selective amplification of restriction fragments by PCR from digested genomic DNA, with restriction site-adapter-specific primers under stringent conditions (39). The restriction fragments analyzed are small, and even mutation of 1 bp can be detected. The use of different sets of restriction enzymes or different primer pair combinations can generate large numbers of different AFLP fingerprints without prior knowledge of sequence. Fluorescent AFLP, which uses fluorescent dye-labeled primer, has proved to be reproducible and capable of standardization (4, 15). It was shown elsewhere to have a discriminatory power equal to that of PFGE in genotyping S. enterica serovars (25) and higher than that of PFGE in genotyping S. enterica serovar Enteritidis phage type 4 (10).
In the present study, fluorescent AFLP was applied to nine phage types of serovar Typhimurium to explore their genetic relationships; to study the genetic variations detected by AFLP, and to assess the potential of using markers identified by AFLP for multiplex PCR or microarray technology as a successor to phage typing.
MATERIALS AND METHODS
Bacterial isolates and DNA preparation.
Forty-six serovar Typhimurium isolates from nine phage types were used. The selected phage types, DT9, DT135, DT64, DT44, DT126, DT12a, DT1, DT141, and DT108, have been dominant or frequent phage types in animal and human infections in Australia in recent years (30, 31). All isolates used in this study were kindly provided by the Institute of Medical and Veterinary Science (IMVS) in Adelaide, Australia. The isolates were from different sources, including human, animal, and environmental sources in different regions of Australia (Table 1). Genomic DNA preparation was as described previously by Bastin et al. (6). Plasmid DNA was extracted by the alkaline method of Kado and Liu (21).
TABLE 1.
Serovar Typhimurium isolates used in this study
| Phage type (DT) | Strain no. | Original ID no.a | Sourceb |
|---|---|---|---|
| 9 | M1844 | 97/05047 | Bovine, NSW |
| M1845 | 97/05933 | Chicken, NSW | |
| M1846 | 97/05027 | Emu intestine, SA | |
| M1847 | 97/05966 | Human, SA | |
| M1848 | 97/04970 | Chicken litter, SA | |
| 135 | M1849 | 97/05944 | Avian, Vic |
| M1850 | 97/06193 | Bovine, NSW | |
| M1851 | 97/05322 | Environment, Vic | |
| M1852 | 97/05570 | Macadamia nuts, NSW | |
| M1853 | 97/03475 | Soya meal, NSW | |
| 64 | M1854 | 97/05010 | Chicken, NSW |
| M1855 | 97/03023 | Bovine, Qld | |
| M1856 | 97/01630 | Human abortion, SA | |
| M1857 | 97/02201 | Porcine lymph node, SA | |
| M1858 | 97/05229 | Human, NSW | |
| 126 | M1859 | 97/05638 | Alpaca, NSW |
| M1860 | 97/06017 | Chicken litter, Qld | |
| M1861 | 97/05747 | Human, SA | |
| M1862 | 97/05741 | Meerkat, NSW | |
| M1863 | 97/04024 | Quail, NSW | |
| M1878 | 98/00437 | Human blood, NSW | |
| 12a | M1864 | 97/03955 | Human, NSW |
| M1865 | 97/04066 | Human abscess, NT | |
| M1866 | 97/03009 | Bovine, NSW | |
| M1867 | 97/02254 | Snake, SA | |
| M1868 | 97/06191 | Feline, SA | |
| M1869 | 97/06333 | Goat meat, SA | |
| M1870 | 97/02501 | Human urine, SA | |
| 44 | M1871 | 97/05455 | Bovine, NSW |
| M1872 | 97/05136 | Human, SA | |
| M1873 | 97/03004 | Human urine, SA | |
| M1874 | 97/01065 | Kangaroo, NSW | |
| M1875 | 98/00592 | Chicken liver, Qld | |
| M1876 | 98/00413 | Ovine intestine, SA | |
| 1 | M1877 | 97/06772 | Human, SA |
| M1879 | 97/03558 | Human, NT | |
| M1880 | 97/06504 | Human, SA | |
| 141 | M1881 | 97/06110 | Bovine, SA |
| M1882 | 97/05604 | Human, SA | |
| M1883 | 97/03996 | Bovine, Qld | |
| M1884 | 97/03253 | Human, NSW | |
| 108 | M1885 | 97/06503 | Bovine lymph node, NSW |
| M1886 | 97/05372 | Beef, Qld | |
| M1887 | 97/04021 | Bovine, SA | |
| M1888 | 97/03007 | Avian, NSW | |
| M1889 | 97/02980 | Ovine, NSW |
Original identification (ID) number is that used by the IMVS in Adelaide, Australia, where these isolates were obtained.
Abbreviations: NSW, New South Wales; SA, South Australia; Vic, Victoria; Qld, Queensland; NT, Northern Territory (all in Australia).
AFLP reaction.
The AFLP technique was performed as described previously by Lan and Reeves (23, 24) for radioactive AFLP, and for fluorescent AFLP the primer was labeled with fluorescent dye.
Genomic DNA was digested by EcoRI and MseI and ligated to EcoRI and MseI adapters simultaneously (note that ligation to adapters does not generate a full restriction site). Fluorescent AFLP was done with 6-carboxyfluorescein (FAM; blue) fluorescent dye-labeled MseI primer (ABI Perkin-Elmer) with one base selection (MseI +1) and unlabeled EcoRI primer with one base selection (EcoRI +1).
Analysis of AFLP fingerprinting patterns.
One microliter of each fluorescent AFLP product was electrophoresed on an ABI 373 DNA sequencer equipped with the ABI PRISM GeneScan 2.0 software at the Sydney University Prince Alfred Macromolecular Analysis Centre. A GeneScan TAMRA-500 internal size standard (Perkin-Elmer) was also loaded with each AFLP sample to enable precise size determination of amplified fragments. The data for each lane were saved as an individual GeneScan file and displayed as an electropherogram: peaks representing AFLP fragments from 40 to 600 bp were visually inspected, and presence was scored as 1 and absence was scored as 0. Most peaks were easily scored for presence or absence, but some low-intensity peaks were judged by comparison with the same peak of other strains. Generally peaks with intensity less than 1/10 of that of the peak with the highest intensity were not scored in this study. Those peaks that were included could be scored as present or absent without ambiguity. The accuracy of scoring was confirmed when the 18 fragments cloned gave the expected pattern when used as a probe for blotting (see Results and Discussion). The Dice coefficient (SD) was used for phylogenetic tree construction with the unweighted pair group method with arithmetic mean (34) in PHYLIP (13).
AFLP was done for all 16 possible MseI +1-EcoRI +1 primer pair combinations. Primer pair MseI + C-EcoRI + A is represented as C/A, etc. The fragments further analyzed were given a number following the primer pair combination. C/T-1 and C/T-2 are polymorphic fragments generated with primer pair MseI + C and EcoRI + T, etc. These numbers do not indicate relative mobility.
Cloning and sequencing of polymorphic AFLP fragments.
Polymorphic AFLP fragments were excised from a 6% polyacrylamide gel after radioactive AFLP, with AFLP patterns visualized by autoradiography for 48 h with Kodak MR BioMax film. The excised fragments were cloned into the pGEM-T Easy plasmid cloning vector (Promega) and transformed into Escherichia coli K-12 strain JM109 by electroporation with a Bio-Rad gene pulser. The recombinant plasmid was extracted from the clone with the Wizard 373 DNA purification system (Promega). For each excised fragment, up to 10 recombinant plasmids were screened for the correct polymorphic AFLP fragment by probing unlabeled AFLP polyacrylamide gel blots with DIG Easy Hyb and the DIG luminescent detection kit for nucleic acids (Boehringer Mannheim) with PCR-amplified insert DNA labeled by the DIG DNA labeling kit (Boehringer Mannheim). In each case one or more plasmids were found with an insert that when used as a probe showed the same polymorphism as observed in fluorescent AFLP and radioactive AFLP. One such insert was sequenced for each of the 18 fragments.
Plasmid sequencing was carried out by the Sydney University Prince Alfred Macromolecular Analysis Centre using an ABI 377A automated DNA sequencing system and the ABI dye terminator cycle sequencing kit (Perkin-Elmer).
Analysis of AFLP fragment sequences.
BlastN and BlastX searches were carried out with ANGIS (Australian National Genomic Information Service), which incorporates several sets of programs (32). The searches were against the nonredundant nucleic and the nonredundant protein databases compiled by ANGIS. BlastN searches were also carried out against the S. enterica serovar Typhimurium, serovar Paratyphi, and serovar Typhi genome sequences at http://genome.wustl.edu/gsc/bacterial/salmonella.shtml and http://www.sanger.ac.uk/Projects/S_typhi/blast_server.shtml. We used the Enterix site, http://galapagos.cse.psu.edu/enterix, for sequences that matched genes in the above genomes. The alignment of the genomes for the above three and also the serovar Dublin and serovar Enteritidis (incomplete) genomes allowed us to determine for each gene if it was present or absent in each of the five genomes.
Southern hybridization.
Plasmid DNA or EcoRI-digested genomic DNA was electrophoresed on an 0.7% agarose gel and transferred to a Hybond-N+ nylon hybridization membrane with a VacuGene XL vacuum blotting system (Amersham Pharmacia Biotech). Cloned AFLP fragments were amplified by PCR, labeled with the DIG DNA labeling kit (Boehringer Mannheim), and hybridized to the blots at high stringency with DIG Easy Hyb and the DIG luminescent detection kit for nucleic acids (Boehringer Mannheim).
Nucleotide sequence accession numbers.
The nucleotide sequences of the cloned and sequenced polymorphic AFLP fragments were deposited with GenBank under accession numbers as follows: C/A-1, AF500153; C/A-2, AF500154; C/A-3, AF500155; C/A-4, AF500156; C/A-5, AF500157; C/A-6, AF500158; C/A-7, AF500159; C/C-1, AF500160; C/C-2, AF500161; C/G-1, AF500162; C/G-2, AF500163; C/T-1, AF500164; C/T-2, AF500165; G/A-1, AF500166; G/C-1, AF500167; G/C-2, AF500168; C/C-4, AF500169; and C/C-5, AF500170.
RESULTS AND DISCUSSION
Distribution of AFLP fragments and phylogenetic tree based on AFLP variation.
Forty-six serovar Typhimurium isolates comprising nine phage types were analyzed by AFLP with all 16 possible MseI +1-EcoRI +1 primer pair combinations. For primer pairs C/A and C/T, AFLP was done twice for all isolates studied to test reproducibility, and all the fragments being scored were reproducible.
A total of 1,340 fragments were scored from the 16 primer pair combinations. There were 345 polymorphic fragments, of which 145 were found in only one isolate, leaving 200 phylogenetically informative fragments (Table 2). The distribution of 84 fragments was very closely correlated with phage type (see below). Thirty-five different AFLP patterns were observed in the 46 isolates. AFLP fingerprints were clearly different between phage types. The Simpson index of diversity (18) was calculated as 0.98.
TABLE 2.
Numbers and types of polymorphisms observed in AFLP fragments in the size range from 40 to 600 bp
| Primer paira | No. of fragments
|
||
|---|---|---|---|
| Total | Polymorphic | Informative | |
| A/A | 92 | 22 | 8 |
| A/C | 84 | 18 | 12 |
| A/G | 94 | 17 | 6 |
| A/T | 103 | 48 | 31 |
| C/A | 106 | 27 | 14 |
| C/C | 111 | 17 | 10 |
| C/G | 114 | 22 | 13 |
| C/T | 65 | 23 | 12 |
| G/A | 54 | 20 | 13 |
| G/C | 52 | 13 | 7 |
| G/G | 54 | 8 | 4 |
| G/T | 41 | 17 | 6 |
| T/A | 87 | 21 | 14 |
| T/C | 83 | 25 | 17 |
| T/G | 113 | 24 | 17 |
| T/T | 87 | 23 | 16 |
| Total | 1,340 | 345 | 200 |
Primer pair combination: MseI + 1-EcoRI + 1.
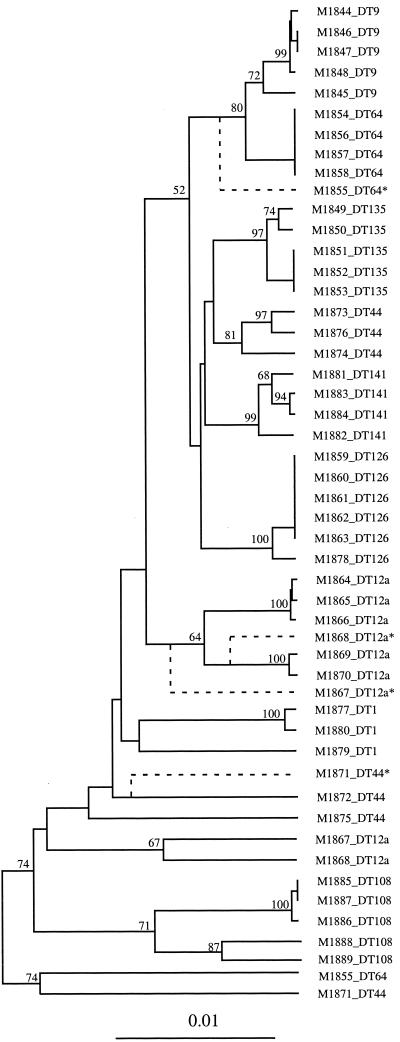
A phylogenetic tree (Fig. 1) was constructed. The tree showed a good correlation with phage type, grouping most isolates of the same phage type together. However, there were exceptions. DT64 isolate M1855 and DT44 isolate M1871 clustered outside the main clusters due to an additional 22 fragments present in these two isolates only. These fragments, most likely of plasmid or phage origin, represent gain of DNA shared by these two isolates. In a tree that was derived after excluding these 22 fragments, M1855 is close to the DT64 and DT9 cluster, and M1871 is close to two other DT44 isolates (Fig. 1 and legend). Even after exclusion of the 22 fragments discussed above, the six DT44 isolates still did not form a single cluster. The seven DT12a isolates fell into two groups. Isolates M1867 and M1868 were clustered together away from the other five DT12a isolates, due to the presence of seven fragments in these two isolates only. Again, in a tree that was derived after excluding these seven fragments, M1867 and M1868 moved into the main DT12a cluster.
FIG. 1.
Phylogenetic tree based on AFLP fingerprints with all 16 primer pair combinations of MseI +1-EcoRI +1 from MseI- and EcoRI-digested fragments. Bootstrap values are percentages of 1,000 replications and are indicated at the nodes if the value is greater than 50%. Isolates indicated by an asterisk are shown twice, in the position found in the tree as originally derived (solid line) and in the position found when fragments specific to these isolates were omitted (dashed line; see text). The value of the bar represents genetic distance measured by Dice coefficient.
Phage-type-correlated AFLP fragments.
AFLP analysis revealed many phage-type-specific markers. There were 59 phage-type-specific polymorphic AFLP fragments, for which presence or absence correlated absolutely with phage type, and 25 for which there was only one exception to phage type specificity in the 46 isolates studied (Table 3 These 84 fragments were analyzed in detail, as they have the potential to be used to subdivide serovar Typhimurium in the same way as achieved by phage typing. Clearly the chance of finding an exception to phage type specificity goes up with the number of isolates studied, and there is no reason to think that the 59 fragments with 100% correlation would be very different in distribution than the 25 others in a much larger sample. In general we treat all 84 together but for some purposes distinguish the 25 that have a single exception to phage type specificity.
TABLE 3.
Phage-type-specific AFLP fragments
| AFLP fragment | Sizeb (bp) | Result for phage type (DT)a:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9 | 12a | 44 | 64 | 108 | 126 | 135 | 141 | ||
| A/A-1 | 343 | + | + | + | + | − | + | + | + | + |
| A/A-2c | 227 | −h | + | + | + | + | + | + | + | + |
| A/C-1 | 182 | + | + | + | + | + | + | − | + | + |
| A/C-2 | 353 | + | − | − | − | − | − | − | − | − |
| A/C-3c | 91 | −h | + | − | + | + | + | + | + | − |
| A/C-4c | 108 | −h | − | − | − | − | + | − | − | − |
| A/C-5c | 229 | +i | − | − | − | − | − | − | − | − |
| A/G-1 | 149 | − | − | − | − | − | − | + | − | − |
| A/G-2 | 150 | + | + | + | + | + | + | − | + | + |
| A/G-3 | 222 | − | − | − | − | − | + | − | − | − |
| A/T-1 | 68 | − | − | + | − | + | + | − | − | − |
| A/T-2 | 110 | + | + | + | + | − | + | + | + | + |
| A/T-3 | 165 | − | − | − | − | − | + | − | − | − |
| A/T-4 | 182 | − | − | − | − | − | − | + | − | − |
| A/T-5 | 454 | − | − | − | − | − | + | − | − | − |
| A/T-6 | 461 | − | − | − | − | − | − | + | − | − |
| A/T-7c | 116 | − | − | − | −j | − | + | − | − | − |
| A/T-8c | 218 | − | − | − | − | + | − | − | −k | − |
| A/T-9c | 415 | − | + | − | +l | + | − | − | − | − |
| A/T-10c | 534 | +i | − | − | − | − | − | − | − | − |
| C/A-1d | 156 | − (−) | − (−) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) | − (−) |
| C/A-2d | 240 | − (−) | − (−) | + (+) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) |
| C/A-3d | 244 | − (−) | − (−) | − (−) | − (−) | − (−) | + (+) | − (−) | + (+) | − (−) |
| C/A-4d | 212 | − (+) | + (+) | − (+) | − (+) | + (+) | − (+) | − (+) | − (+) | − (+) |
| C/A-5d | 208 | + (+) | − (+) | + (+) | + (+) | − (+) | + (+) | + (+) | + (+) | + (+) |
| C/A-6d | 69 | − (−) | − (−) | + (+) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) |
| C/A-7d | 122 | + (+) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−e) |
| C/A-8 | 305 | − | − | − | − | − | − | + | − | − |
| C/C-1d | 171 | + (+) | + (+) | + (+) | + (+) | + (+) | + (+) | − (+) | + (+) | + (+) |
| C/C-2d | 179 | − (+) | − (+) | − (+) | − (+) | − (+) | − (+) | + (+) | − (+) | − (+) |
| C/C-3 | 276 | − | + | − | − | + | − | − | − | − |
| C/C-4c,d | 136 | + (+) | + (+) | +m (+)f | + (+) | + (+) | + (+) | + (+) | + (+) | − (−e) |
| C/C-5c,d | 313 | + (+) | + (+) | +m (+)f | + (+) | + (+) | − (+) | + (+) | + (+) | − (−m) |
| C/C-7c | 63 | − | − | − | −j | − | + | − | − | − |
| C/C-8c | 106 | +i | − | − | − | − | − | − | − | − |
| C/G-1d | 218 | − (−) | − (−) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) | − (−) |
| C/G-2d | 121 | + (+) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−e) |
| C/G-3 | 414 | − | + | − | − | + | − | − | − | − |
| C/G-4 | 311 | − | − | − | − | + | − | − | − | − |
| C/G-5 | 85 | − | − | − | − | − | + | − | − | − |
| C/G-6c | 136 | − | − | − | − | − | +n | − | − | − |
| C/T-1d | 175 | − (−g) | − (−) | + (+) | − (−) | + (+) | + (+) | − (−) | − (−) | − (−) |
| C/T-2d | 275 | + (+) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−) | − (−e) |
| C/T-3 | 129 | − | − | − | − | − | + | − | − | − |
| C/T-4c | 441 | −h | + | − | + | + | + | + | + | − |
| G/A-1d | 132 | − (−) | − (−) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) | − (−) |
| G/A-2 | 291 | + | − | − | − | − | − | − | − | − |
| G/A-3 | 347 | + | + | + | + | + | + | − | + | + |
| G/A-4c | 114 | +i | − | − | − | − | − | − | − | − |
| G/A-5c | 280 | − | − | − | +l | − | − | − | − | − |
| G/C-1d | 182 | − (−) | − (−) | + (+) | − (−) | − (−) | + (+) | − (−) | − (−) | − (−) |
| G/C-2d | 200 | − (+) | − (+) | − (+)f | − (+) | − (+) | + (+) | − (+) | − (+) | − (−e) |
| G/C-3 | 474 | − | − | − | − | − | + | − | − | − |
| G/C-4 | 598 | + | − | − | − | − | − | − | − | − |
| G/T-1c | 47 | + | − | − | − | − | − | −o | − | − |
| G/T-2c | 525 | +i | − | − | − | − | − | − | − | − |
| T/A-1 | 94 | − | − | − | − | − | + | − | − | − |
| T/A-2 | 127 | + | − | − | − | − | − | − | − | − |
| T/A-3 | 162 | + | − | − | − | − | − | − | − | − |
| T/A-4 | 170 | + | − | − | − | − | − | − | − | − |
| T/A-5 | 216 | − | − | − | − | − | + | − | + | − |
| T/A-6 | 376 | − | + | − | − | + | − | − | − | − |
| T/C-1 | 96 | + | − | − | − | − | − | − | − | − |
| T/C-2c | 198 | +i | − | − | − | − | − | − | − | − |
| T/G-1 | 48 | + | − | − | − | − | − | − | − | − |
| T/G-2 | 206 | − | − | + | − | − | + | − | − | −/PICK> |
| T/G-3 | 375 | − | + | − | − | + | − | − | − | − |
| T/G-4 | 415 | − | − | − | − | + | − | − | − | − |
| T/G-5c | 56 | + | − | − | − | − | − | −o | − | − |
| T/G-6c | 75 | + | − | − | − | − | − | −o | − | − |
| T/G-7c | 80 | −h | + | + | + | + | + | + | + | + |
| T/G-8c | 394 | + | − | + | + | −p | + | + | + | + |
| T/T-1 | 67 | − | − | − | − | − | − | + | − | − |
| T/T-2 | 92 | − | − | − | − | + | − | − | − | − |
| T/T-3 | 185 | − | − | + | − | − | + | − | − | − |
| T/T-4 | 233 | − | − | − | − | − | − | + | − | − |
| T/T-5 | 234 | + | + | + | + | + | + | − | + | + |
| T/T-6 | 241 | − | − | + | − | − | − | − | − | − |
| T/T-7 | 278 | − | − | − | − | − | + | − | − | − |
| T/T-8 | 478 | − | − | − | − | − | + | − | − | − |
| T/T-9 | 550 | − | − | − | − | − | + | − | − | − |
| T/T-10 | 572 | − | − | − | − | − | + | − | − | − |
| T/T-11c | 104 | − | −q | − | − | − | + | − | + | − |
| T/T-12c | 454 | +i | − | − | − | − | − | − | − | − |
Hybridization of cloned phage-type-specific AFLP fragments to EcoRI-digested genomic DNA of the serovar Typhimurium isolates studied is shown in parentheses.
Sizes are precise for the fragments that were sequenced.
Fragment with a single exception to phage type specificity.
Fragment cloned and sequenced.
Hybridized to isolate M1882 at a different size at low stringency.
Did not hybridize to isolate M1867.
Hybridized to several different-sized fragments in isolate M1879 at low stringency.
Present in isolate M1879.
Absent in isolate M1879.
Present in isolate M1875.
Present in isolate M1850.
Absent in isolate M1875.
Absent in isolate M1867.
Absent in isolate M1886.
Present in isolate M1878.
Present in isolate M1855.
Present in isolate M1848.
Of the 84 phage-type-specific fragments, if we ignore the single exception for the 25 fragments, 63 are present only or absent only in one phage type. Five are present only and two are absent only in DT9 and DT64, five are present only in DT12a and DT108, three are present only in DT108 and DT135, and one is absent only in DT108 and DT141. Two are present only in DT12a, DT64, and DT108. Two are absent only in DT1, DT12a, and DT141, and one is present only in DT9, DT44, and DT64. Of the exceptions to phage type specificity in the 25 near-specific fragments, 12 involve DT1 isolate M1879; four involve DT44 isolate M1875; three involve DT126 isolate M1878; two involve DT12a isolate M1867; and single fragments involve each of the isolates DT64 isolate M1855, DT108 isolate M1886, DT135 isolate M1850, and DT9 isolate M1848. The nine phage types studied could all be differentiated by the 84 fragments.
Nature of phage-type-correlated AFLP fragments.
Eighteen phage-type-specific fragments were cloned and sequenced to determine the genetic basis of AFLP-detected variation. The sequences were used in BlastN and BlastX searches (Table 4). The 18 fragments were also used to probe EcoRI-digested genomic DNA of the 46 isolates (Table 3). Fifteen have high sequence similarity to known genes or sequences in databases. Three matched plasmid genes, seven matched lambdoid phage genes, one matched a noncoding region of a lambdoid phage genome, and four matched chromosomal genes or sequences in the available S. enterica genomes.
TABLE 4.
Sequences of phage-type-specific AFLP fragments
| Fragment | Size (bp) | Identitya (%) | Gene(s) | Accession no. | Function | Similarity region |
|---|---|---|---|---|---|---|
| C/A-1 | 156 | 97 | Bacteriophage, P22 eac gene | L06296 and AF217253 | EaC protein | P22 genome 4918-5049 |
| C/A-2 | 240 | No match | ||||
| C/A-3 | 244 | No match | ||||
| C/A-4b | 212 | 98 | S. enterica genome sequence | LT2: AE006468 | Serovar Typhimurium LT2 1096650-1096833 | |
| C/A-5b | 208 | 100 | S. enterica genome sequence | LT2: AE006468 | Serovar Typhimurium LT2 1096650-1096833 | |
| C/A-6 | 69 | 100 | Bacteriophage 21 DNA | AJ237660 | Noncoding region | 4846-4883 |
| C/A-7 | 122 | 100 | Plasmid R64 traQ gene | AB027308 | 33387-33484 | |
| C/C-1c | 171 | 100 | S. enterica genome sequence | LT2: AE006468 | Serovar Typhimurium LT2 1766735-1766881 | |
| C/C-2c | 179 | 98 | S. enterica genome sequence | LT2: AE006468 | Serovar Typhimurium LT2 1766735-1766881 | |
| C/G-1 | 218 | 31 (BlastX) | Bacteriophage P22 gene 16 | M74136 | DNA transfer protein GP16 | |
| C/G-2 | 121 | 97 | Plasmid R64 trbC gene | AB027308 | Nucleotide-binding protein | 50509-50608 |
| C/T-1 | 175 | 100 | Bacteriophage P22 gene 8 | M59749 and AF217253 | Scaffold protein GP8 | P22 genome 27405-27555 |
| C/T-2 | 275 | 100 | Plasmid R64 traP and traQ genes | AB027308 | 33140-33390 | |
| G/A-1 | 132 | 75 (BlastX) | Bacteriophage HK97 gene 38 | AF069529 | Protein GP38 | |
| 66 (BlastX) | Bacteriophage P22 ORF-56 | AAF75011 | ||||
| G/C-1 | 182 | No match | ||||
| G/C-2 | 200 | 94 | ORF626 of S. enterica serovar Dublin contig 707 | 40-213 | ||
| C/C-4 | 136 | 95 | ORF626 of S. enterica serovar Dublin contig 707 | 210-321 | ||
| C/C-5 | 313 | 96 | ORF626 of S. enterica serovar Dublin contig 707 | 1-213 |
BlastN unless otherwise specified.
C/A-4 and C/A-5 are alternative fragments with high-level identity to the same S. enterica genome sequence.
C/C-1 and C/C-2 are alternative fragments with high-level identity to the same S. enterica genome sequence.
Fragments with genes of plasmid origin.
Three fragments (C/G-2, C/T-2, and C/A-7, all present only in DT1) have 97 to 100% DNA sequence identity to plasmid R64 trbC, traP, and traQ genes (Table 4) (22). The latter two matched contiguous sequences, being bases 33140 to 33390 and bases 33387 to 33484 of plasmid R64, respectively, connected by an EcoRI site. These three fragments hybridized to large plasmid DNA (data not shown) present in DT1 isolates only. It seems clear that DT1 isolates have a large plasmid not present in the 43 other isolates.
Fragments with genes of lambdoid phage origin.
Three fragments (C/A-1, C/G-1, and G/A-1), all present in DT64 only, also hybridized to DT64 only. Their sequences matched those of lambdoid phage genes. C/A-1 has 97% sequence identity to the lambdoid phage P22 DNA at positions 4918 to 5049 (37) on the eac gene (42). C/G-1 has 31% sequence identity at the amino acid level to phage P22 gene 16, which encodes a DNA transfer protein (36). G/A-1 has 75% sequence identity at the amino acid level to lambdoid phage HK97 gene 38 (20). G/A-1 also has 66% sequence identity at the amino acid level to phage P22 ORF-56, a hypothetical 6.6-kDa protein in the eae-abc2 intergenic region (37). It seems clear that DT64 carried a lambdoid phage not present in the other eight phage types.
Fragment C/T-1 is shared by DT64, DT12a, and DT108 isolates and also hybridized to DT64, DT12a, and DT108 only. C/T-1 has 100% sequence identity to the phage P22 DNA at positions 27405 to 27555 (37) on gene 8, which encodes scaffold protein (12). Fragment C/A-6, present only in DT12a and DT108, also hybridized to DT12a and DT108 only. It has 100% sequence identity to a noncoding region of the lambdoid phage 21 genome at positions 4846 to 4883. The above data indicated the presence of lambdoid phages in DT64, DT12a, and DT108. It also indicated that some genes are common to these phages.
Three fragments (G/C-2, C/C-4, and C/C-5) have 94 to 96% DNA sequence identity to DNA of ORF626 of S. enterica serovar Dublin contig 707, and ORF626 has 40% sequence identity at the amino acid level to the phage P1 kilA gene. KilA protein is nonessential for phage replication and lytic development (17). G/C-2 and C/C-5 both are contiguous with C/C-4 across an EcoRI site. G/C-2, C/C-5, and C/C-4 matched bases 40 to 213, bases 1 to 213, and bases 210 to 321 of ORF626, respectively. The G/C-2 sequence has 97% identity to part of the C/C-5 sequence with an EcoRI site at the junction. The C/C-5 sequence has an additional segment extending from a 5′-TTGA-3′ site in place of the 5′-TTAA-3′ MseI site of G/C-2. The distribution of the fragments showed that DT12a isolate M1867 and DT141 have none of these three fragments and also did not hybridize with them. All other isolates except DT108 have fragments C/C-4 and C/C-5, while DT108 isolates have fragments C/C-4 and G/C-2. The above data suggested that both C/C-4 and C/C-5 are from a phage-related DNA sequence and that fragment G/C-2 arose through a point mutation that created a MseI site in DT108, generating an AFLP fragment shorter than C/C-5.
Four fragments related by mutational change adjacent to chromosomal genes.
Two of the sequenced fragments (C/A-4 and C/A-5) are inversely correlated. C/A-4 is present only in DT9 and DT64, while C/A-5 is absent only in these phage types. The sequences are nearly identical, and the C/A-5 sequence was found in the five publicly available sequences of S. enterica genomes (serovars Typhimurium, Paratyphi, Typhi, Enteritidis, and Dublin). The C/A-5 sequence is 100% identical to serovar Typhimurium LT2 DNA bases 1096650 to 1096833. There is a four-base duplication in C/A-4, relative to C/A-5, containing 5′-GGTAGGTA-3′ and 5′-GGTA-3′, respectively, in an intergenic region between STM1003 and the pncB gene (nicotinate phosphoribosyltransferase [38]) at serovar Typhimurium LT2 genome sequence bases 1096771 to 1096774 (26). pncB is a housekeeping gene and also present in other bacteria, such as E. coli, Klebsiella pneumoniae, Yersinia pestis, and Vibrio cholerae, etc., according to the Enterix site.
Another two fragments (C/C-1 and C/C-2) are also inversely correlated. C/C-1 is absent only in DT126, while C/C-2 is present only in DT126. The sequences are nearly identical, and the C/C-1 sequence was found in four of the publicly available sequences of S. enterica genomes (serovars Typhimurium, Paratyphi, Typhi, and Enteritidis) but absent in serovar Dublin. The C/C-1 sequence is 100% identical to serovar Typhimurium LT2 DNA sequence bases 1766735 to 1766881. There is an eight-base duplication in C/C-2 relative to C/C-1, containing 5′-ACAGACCGACAGACCG-3′ and 5′-ACAGACCG-3′, respectively, in an intergenic region between STM1672 and STM1673 at serovar Typhimurium LT2 genome sequence bases 1766792 to 1766799 (26).
For both pairs of fragments, the larger fragment is in one or two serovar Typhimurium DTs only, while the smaller fragment is present in most serovar Typhimurium DTs and the other three or four sequenced S. enterica genomes in which the gene is found. It seems clear that the mutations were due to duplication of a short sequence and not vice versa.
Fragments with genes of unknown origin.
Three phage-type-specific fragments (C/A-2, G/C-1, and C/A-3) did not have sequence similarity with any DNA or protein entry in the database searches. C/A-2 and G/C-1 are present in DT12a and DT108 only and also hybridized to DT12a and DT108 only. C/A-3 is present in DT135 and DT108 only and also hybridized to DT135 and DT108 only. These fragments were most likely polymorphic due to gain of DNA.
Predominance of phage and plasmid genes in phage-type-related AFLP fragments.
Of the 18 AFLP fragments cloned and sequenced, 15 comprise or include genes identifiable to some extent. Three are plasmid related, eight are phage related, and four are chromosomal gene related.
Six of the fragments comprise three pairs that differ in one or a few base pairs. The pair C/A-4 and C/A-5, the pair C/C-1 and C/C-2 (each pair comprising different forms of the same intergenic region), and the pair C/C-5 and G/C-2 differ by a base substitution that affects an MseI site in a phage-related gene. These six fragments thus correspond to three loci, each with two alleles, giving a total of 15 AFLP-based loci for the 18 fragments. Two of these mutational changes are in intergenic regions adjacent to chromosomal genes.
Hybridization at high stringency showed that the six fragments discussed above comprise DNA generally present in all nine phage types, the only exception being the phage-related gene which was absent in DT141 and DT12a isolate M1867. However, the other 12 fragments contained DNA present only in isolates with that fragment. This included all of the phage-related genes and plasmid-related genes. These results suggested that gain or loss of DNA rather than mutational change was responsible for most of the polymorphisms detected by AFLP. It should be noted that our focus has been on fragments that correlated strongly with phage types and that this generalization may not apply to all polymorphic fragments.
However, some cross hybridization was observed at low stringency (Table 3). C/T-1 gave low-level hybridization to several fragments of different sizes from DT1 isolate M1879, suggesting that M1879 carried a phage related to that in DT12a, DT64, and DT108. Three plasmid R64-related fragments (C/G-2, C/T-2, and C/A-7, present only in DT1) and three phage P1-related fragments (C/C-5, G/C-2, and C/C-4) also gave low-level hybridization to one fragment from DT141 isolate M1882. These cases of cross hybridization indicated the presence of related DNA, perhaps on a plasmid related to that in DT1 or on a phage related to P1.
The data overall indicated that the major consistent differences between phage types relate to the presence or absence of phages or plasmids, well known as mobile genetic elements. The data are certainly consistent with earlier observations showing the role of plasmids and phages in alteration or determination of phage types (2, 41). For example, a study of DT49 and DT204 isolates of human and bovine origin in Britain showed a correlation among plasmid content, antibiotic resistance spectra, and phage type (41). Tetracycline-resistant DT204 strains all carried nonautotransferring plasmids coding for tetracycline resistance only. Spontaneous loss of tetracycline resistance plasmid NTP108 converted a DT204 strain to a DT49 strain. When plasmid NTP108 was reintroduced into the DT49 strain, the phage type changed back to DT204. DT204 differed from DT49 by loss or reduction in sensitivity to specific typing phages, and further experiments showed that plasmid NTP108 restricted growth of these specific serovar Typhimurium typing phages (41).
The identification of phage-type-specific phage and plasmid genes provides a basis for exploration of the genetic basis of phage resistance and phage type variation. However, our data do not address the relationship of the phages and plasmids detected with the various sensitivities of each phage type to the typing phages. It is possible that some of the phages or plasmids whose presence was detected in this study are simply markers for variants of serovar Typhimurium. The fact that phage and plasmid genes were dominant in our observations does not in itself imply that these elements affect sensitivity to typing phages but suggests only that the rate of gain and loss of these genetic elements affects AFLP pattern more rapidly than gain or loss of restriction sites by mutation or lateral transfer of chromosomal genes. This is an interesting topic for further study. While this paper was being written, Tamada et al. (35) reported sequences of two AFLP markers of serovar Typhimurium. One sequence showed homology with a segment of P22 phage, and the other showed homology with a segment of traG, an F-plasmid conjugation gene. These data are consistent with our data.
Given the role of phage-related genes in our AFLP tree and the role attributed to carriage of temperate phage in determining phage type, we compared the relationships of phage types revealed by the AFLP tree with the relationships based on the phage sensitivity in the Anderson phage-typing scheme. The sensitivity of each isolate to each typing phage is assessed by number and turbidity of plaques (3, 9). The lysis pattern for each of 30 phages as updated (Linda Ward, personal communication) was used to grade the sensitivity of each of the nine DTs in steps from 1 to 24 for increasing degree of lysis. These data were treated as 30 loci each with 24 character steps for construction of the tree, such that relationship would be determined by degree of similarity based on the 24-step scale, and a phenotypic tree was constructed by the parsimony method (data not shown). The phage sensitivity tree was very different from the AFLP tree, with, for example, DT12a, DT64, and DT108 closely related in the phage sensitivity tree but not in the AFLP tree. We suggest that the relationships in the AFLP tree are determined to a significant degree by the overlap in genes present in phages and plasmids carried by the strains, whereas the phage sensitivity tree is determined by the degree of sensitivity to specific typing phages, which are apparently not closely related.
Comparison of AFLP with PFGE.
Several molecular methods have been tested for use in subtyping of common S. enterica serovars such as Typhimurium. The only one that has achieved a significant level of use is PFGE, which in some cases has also been used in place of phage typing for primary subdivision of serovar Typhimurium. It has also proved to be useful in subdividing a major phage type (11). We have studied AFLP in large part because the fragments obtained are small and quite easily cloned and sequenced to determine the underlying basis of the distinctions observed. In the present study, AFLP based on 16 primer pair combinations showed a Simpson index of diversity of 0.98, a higher discriminatory power than that for PFGE applied to Spanish serovar Typhimurium isolates with a Simpson index of diversity of 0.87 (16). AFLP in the present study showed good correlation with phage type, grouping most of the serovar Typhimurium isolates by phage type and enabling differentiation of phage types. However, AFLP is not suitable for routine use for subdivision of a S. enterica phage type, because with such closely related strains one has to use several primer pairs to obtain the high discriminatory power possible, and we still have difficulty with automatic computer scoring of AFLP fragments.
However, this study has demonstrated that AFLP can be used to identify segments of DNA that have a distribution that correlates with phage type. We propose that this knowledge be used to establish a subtyping scheme for serovar Typhimurium that offers the advantages of AFLP with potential for great ease of operation as discussed below.
PFGE also showed some correlation with phage type when applied to Danish serovar Typhimurium isolates (28). However, PFGE uses one restriction enzyme, which cleaves infrequently and generates a small number of large DNA fragments. It can detect mutational changes only in the small number of restriction enzyme sites, or changes such as large insertion-deletion events or chromosomal rearrangements, which cause several-kilobase differences in fragment size. It is difficult to detect insertion-deletion events involving only a few kilobases. In contrast, AFLP uses two restriction enzymes and generates small fragments. AFLP is ideal for detecting insertion or loss of segments of DNA, and it is easy to clone such DNA and determine its distribution among phage types as done in this study. Also, such DNA can be used to develop multiplex PCR or microarray typing. AFLP has more potential than PFGE to discriminate among closely related isolates, as the number of AFLP fragments can be greatly increased by using different enzymes or primer sets, to find more phage-type-specific markers.
Potential for microarray typing of serovar Typhimurium using AFLP markers.
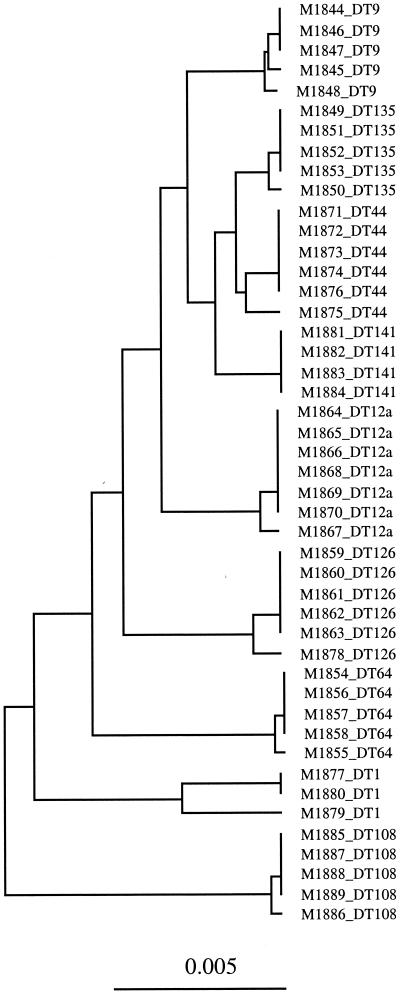
The use of AFLP has enabled us to identify segments of DNA for which presence or absence correlates with phage type. Two-thirds (12 of 18) of cloned phage-type-specific AFLP fragments were polymorphic, because the DNA is present or absent as indicated by Southern hybridization. These AFLP markers provide a good basis for PCR-based typing or application of microarray technology. A tree was constructed by using only the 84 phage-type-specific AFLP fragments (Fig. 2). All nine phage types studied were differentiated, and furthermore, a minimum of six such fragments (C/A-3, C/A-4, C/A-7, C/C-2, C/C-4, and C/T-1) is sufficient to allocate all isolates to their respective phage types. One can envisage the use of multiplex PCR or microarray technology to obtain comparable data for typing, and we suggest that it is entirely practicable to develop subtyping of serovar Typhimurium by using AFLP-defined markers.
FIG. 2.
Tree constructed by using phage-type-specific AFLP fragments of all 16 primer pair combinations of MseI +1-EcoRI +1 from MseI- and EcoRI-digested fragments. The value of the bar represents genetic distance measured by Dice coefficient.
We draw attention to the advantages of AFLP and multiplex PCR for the extensive preliminary work that is needed and for ongoing development of such a scheme. Fluorescent AFLP permits very accurate size standardization of AFLP fragments, which would allow interlaboratory comparisons for the choice of fragments to be used as the basis for a typing scheme. Multiplex PCR using primers based on selected phage-type-specific AFLP fragments would be ideal for trials of such a typing scheme, which could be run in parallel with phage typing. However, microarray technology is better in handling the number of sequences required.
We have looked at only nine phage types and a limited number of isolates from one region of the world. Further work is needed to establish the generality of these observations and to find DNA that gives good correlation with phage typing. We see great advantage in using a scheme that correlates well with phage typing, as the same classification could be followed and the currently named DTs would continue to be recognized.
A microarray-based scheme based on DNA fragments shown to correlate with phage type offers major additional advantages. Unlike PFGE or AFLP, it is relatively easy to scale up to include more test sequences with microarray typing. We have already demonstrated the feasibility of a molecularly based replacement for serotyping (40), and given the huge number of spots that can be fitted onto a microarray, it would be possible to include a full serotyping scheme and several subtyping schemes on one chip. There is the additional advantage that it would be relatively easy to subdivide the frequently occurring DTs, as there is more information available than needed for replacement of phage typing. In this context we should note that phage typing uses a standard system with 16 phages per plate, making scale-up difficult.
The phage-typing system of serovar Typhimurium has proved to be very useful for surveillance of infections. However, the present phage-typing scheme requires considerable experience for consistent scoring and to access specific typing phages. It is difficult to propagate the typing phages to obtain stocks with the same characteristics as the original stocks. This is probably due to recombination between the propagating phage and other phages present in the propagating strain, giving a genetically mixed population in the final stock. For this reason the stocks are prepared in the Enteric Reference Laboratory at the Public Health Laboratory Service in London, United Kingdom, for worldwide distribution. It has even been suggested by Schmieger (33) that, when the stocks of original typing phages become exhausted, the new propagated typing phages may affect the consistency of the Anderson phage-typing scheme.
Microarray technology has much potential for screening with DNA sequence markers. AFLP provides the means to identify suitable DNA segments for use in such technology to provide a tool for subtyping serovar Typhimurium which could be applied more widely than the present phage-typing system, which is generally confined to major government laboratories. This would have a very positive impact on epidemiological investigation of disease outbreaks and studies of pathogenicity of this important pathogen.
Thus, AFLP's real potential for genotyping serovar Typhimurium probably lies in identification of DNA segments that are discriminatory among phage types, followed by multiplex PCR to confirm their value in parallel with phage typing, and then development of microarray technology for routine typing.
Acknowledgments
This research was supported by a grant from the National Health and Medical Research Council of Australia.
We are grateful to Chris Murray of the IMVS, Adelaide, South Australia, Australia, for kindly providing strains. We thank the reviewers for helpful suggestions.
REFERENCES
- 1.Aarts, H. J., L. A. van Lith, and J. Keijer. 1998. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett. Appl. Microbiol. 26:131-135. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. S., E. J. Threlfall, J. M. Carr, M. M. McConnell, and H. R. Smith. 1977. Clonal distribution of resistance plasmid-carrying Salmonella typhimurium, mainly in the Middle East. J. Hyg. 79:425-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, E. S., L. R. Ward, M. J. de Sexe, and J. D. H. de Sa. 1977. Bacteriophage typing designations of Salmonella typhimurium. J. Hyg. 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, C., L. Metherell, G. Willshaw, A. Maggs, and J. Stanley. 1999. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J. Clin. Microbiol. 37:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggesen, D. L., M. N. Skov, D. J. Brown, and M. Bisgaard. 1997. Separation of Salmonella Typhimurium DT2 and DT135: molecular characterization of isolates of avian origin. Eur. J. Epidemiol. 13:347-352. [DOI] [PubMed] [Google Scholar]
- 6.Bastin, D. A., L. K. Romana, and P. R. Reeves. 1991. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol. Microbiol. 5:2223-2231. [DOI] [PubMed] [Google Scholar]
- 7.Bell, C., and A. Kyriakides. 2002. Salmonella—a practical approach to the organism and its control in foods, p. 13. Blackwell Science Ltd., London, United Kingdom.
- 8.Brown, D. J., D. L. Baggesen, D. J. Platt, and J. E. Olsen. 1999. Phage type conversion in Salmonella enterica serotype Enteritidis caused by the introduction of a resistance plasmid of incompatibility group X (IncX). Epidemiol. Infect. 122:19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callow, B. R. 1959. A new typing scheme for Salmonella typhimurium. J. Hyg. 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, M., E. J. Threlfall, and J. Stanley. 2001. Fluorescent amplified-fragment length polymorphism subtyping of the Salmonella enterica serovar Enteritidis phage type 4 clone complex. J. Clin. Microbiol. 39:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebner, P. D., and A. G. Mathew. 2001. Three molecular methods to identify Salmonella enterica serotype Typhimurium DT104: PCR fingerprinting, multiplex PCR and rapid PFGE. FEMS Microbiol. Lett. 205:25-29. [DOI] [PubMed] [Google Scholar]
- 12.Eppler, K., E. Wyckoff, J. Goates, R. Parr, and S. Casjens. 1991. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology 183:519-538. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 14.Gibson, J. R., E. Slater, J. Xerry, D. S. Tompkins, and R. J. Owen. 1998. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J. Clin. Microbiol. 36:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulding, J. N., J. Stanley, N. Saunders, and C. Arnold. 2000. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerra, B., P. Schrors, and M. C. Mendoza. 2000. Application of PFGE performed with XbaI to an epidemiological and phylogenetic study of Salmonella serotype Typhimurium. Relations between genetic types and phage types. New Microbiol. 23:11-20. [PubMed] [Google Scholar]
- 17.Hansen, E. B. 1989. Structure and regulation of the lytic replicon of phage P1. J. Mol. Biol. 207:135-149.2661831 [Google Scholar]
- 18.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 20.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 21.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komano, T., S. Takabe, and H. Sakai. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol. Microbiol. 35:1348-1359. [DOI] [PubMed] [Google Scholar]
- 23.Lan, R., and P. R. Reeves. 2000. Unique adaptor design for AFLP fingerprinting. BioTechniques 29:745-746, 748, 750. [DOI] [PubMed]
- 24.Lan, R., and P. R. Reeves. 2002. Pandemic spread of cholera: genetic diversity and relationships within the seventh pandemic clone of Vibrio cholerae determined by amplified fragment length polymorphism. J. Clin. Microbiol. 40:172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstedt, B. A., E. Heir, T. Vardund, and G. Kapperud. 2000. Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J. Clin. Microbiol. 38:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Y. Du, S. F. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 27.Olsen, J. E., M. N. Skov, O. Angen, E. J. Threlfall, and M. Bisgaad. 1997. Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype typhimurium defined by ribotyping, IS200 typing and PFGE. Microbiology 143:1471-1479. [DOI] [PubMed] [Google Scholar]
- 28.On, S. L. W., and D. L. Baggesen. 1997. Determination of clonal relationships of Salmonella typhimurium by numerical analysis of macrorestriction profiles. Appl. Microbiol. 83:699-706. [DOI] [PubMed] [Google Scholar]
- 29.Popoff, M. Y., and L. L. Minor. 1997. Antigenic formulas of the Salmonella serovars, 7th revision. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 30.Powling, J. (ed.). 1997. National Enteric Pathogens Surveillance Scheme. Human annual report 1996. Microbiological Diagnostic Unit, The University of Melbourne, Melbourne, Victoria, Australia.
- 31.Powling, J. (ed.). 1997. National Enteric Pathogens Surveillance Scheme. Non-human annual report 1996. Microbiological Diagnostic Unit, The University of Melbourne, Melbourne, Victoria, Australia.
- 32.Reisner, A. H., C. A. Bucholtz, J. Smelt, and S. McNeil. 1993. Australia's national genomic information service, p. 595-602. In Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science.
- 33.Schmieger, H. 1999. Molecular survey of Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 35.Tamada, Y., Y. Nakaoka, K. Nishimori, A. Doi, T. Kumaki, N. Uemura, K. Tanaka, S. Makino, T. Sameshima, M. Akiba, M. Nakazawa, and I. Uchida. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umlauf, B., and B. Dreiseikelmann. 1992. Cloning, sequencing, and overexpression of gene 16 of Salmonella bacteriophage P22. Virology 188:495-501. [DOI] [PubMed] [Google Scholar]
- 37.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinitsky, A., H. Teng, and C. T. Grubmeyer. 1991. Cloning and nucleic acid sequence of the Salmonella typhimurium pncB gene and structure of nicotinate phosphoribosyltransferase. J. Bacteriol. 173:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, L., D. Rothemund, H. Curd, and P. R. Reeves. 2000. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J. Clin. Microbiol. 38:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willshaw, G. A., E. J. Threlfall, L. R. Ward, A. S. Ashley, and B. Rowe. 1980. Plasmid studies of drug-resistant epidemic strains of Salmonella typhimurium belonging to phage types 204 and 193. J. Antimicrob. Chemother. 6:763-773. [DOI] [PubMed] [Google Scholar]
- 42.Youderian, P., and M. M. Susskind. 1980. Bacteriophage P22 proteins specified by the region between genes 9 and erf. Virology 107:270-282. [DOI] [PubMed] [Google Scholar]