Abstract
Streptococcus suis is an important pathogen of pigs and occasionally causes serious human disease. However, little is known about the S. suis population structure, the clonal relationships between strains, the potential of particular clones to cause disease, and the relevance of serotype as a marker for epidemiology. Here we describe a multilocus sequence typing (MLST) scheme for S. suis developed in order to begin to address these issues. Seven housekeeping gene fragments from each of 294 S. suis isolates obtained from various S. suis diseases and from asymptomatic carriage representing 28 serotypes and nine distinct countries of origin were sequenced. Between 32 and 46 alleles per locus were identified, giving the ability to distinguish >1.6 × 1011 sequence types (STs). However only 92 STs were identified in this study. Of the 92 STs 18 contained multiple isolates, the most common of which, ST1, was identified on 141 occasions from six countries. Assignment of the STs to lineages resulted in 37 being identified as unique and unrelated STs while the remaining 55 were assigned to 10 complexes. ST complexes ST1, ST27, and ST87 dominate the population; while the ST1 complex was strongly associated with isolates from septicemia, meningitis, and arthritis, the ST87 and ST27 complexes were found to contain significantly higher numbers of lung isolates. In agreement with the observed distribution of disease-causing isolates of S. suis, most isolates previously characterized as of high virulence in porcine infection models belong to ST1, while isolates belonging to other STs appear to be less virulent in general. Finally nine STs were found to contain isolates of multiple serotypes, and many isolates belonging to the same serotypes were found to have very disparate genetic backgrounds. As well as highlighting that the serotype can often be a poor indicator of genetic relatedness between S. suis isolates, these findings suggest that capsular genes may be moving horizontally through the S. suis population.
Streptococcus suis is an important pathogen associated with a range of diseases in pigs, including pneumonia, meningitis, septicemia, and arthritis, although the organism is also frequently carried asymptomatically. S. suis has substantial implications for the swine industry both in terms of animal welfare concerns and economic considerations and can cause serious zoonotic infection of humans, where it has been associated with septicemia, meningitis, and endocarditis (2, 46). The prevention and control of S. suis infection of swine by vaccine and treatment with antimicrobials are hampered by increasing antibiotic resistance and poor understanding of the biology, crucial virulence factors, and protective antigens of the organism (46).
There are currently 35 serotypes of S. suis recognized based on the immunogenicity of capsular antigens (18, 19, 20, 24, 35). Although the majority of disease is caused by a small number of serotypes, it is recognized that the capsular serotype is a poor marker of virulence. Virulence can vary substantially both within and among serotypes, and not all isolates of the same serotype cause the same disease (46). Serotype 2 is considered the most virulent and the most frequently isolated from disease. However, the importance of particular serotypes can vary geographically. For example, some 70% of isolates from porcine disease in France belong to serotype 2 (4), and this serotype was recently reported to be the most frequently isolated in countries such as Italy and Spain (49, 54). In contrast serotype 9 is reportedly particularly common in Belgium and Holland (54) and previously Australia (16), whereas serotypes 1 and 14, in addition to 2, are common in the United Kingdom (22, 54). The prevalence of serotypes also varies temporally. In Canada over the last 7 years the percentage of serotype 2 strains isolated from diseased animals decreased from 22 to 15% (23). Similarly, the importance of serotype 14, which was a major invasive pathogen in the United Kingdom, though apparently not elsewhere, in the middle-to-late 1990s (22), has declined recently.
Previous studies have indicated that S. suis is a genetically diverse species (3, 21, 31, 34, 42) although many involved small numbers of isolates, focused on isolates representing a single serotype or geographical location, and used a variety of traditional approaches such that it is difficult to compare results among studies. In spite of acknowledged difficulties in the categorization of S. suis isolates as virulent or avirulent (see reference 17 for a discussion), there is considerable evidence that virulent isolates are genetically distinct from avirulent isolates, suggesting a clonal association with virulence in S. suis. Staats et al. (47) found that most serotype 2 isolates from septicemia represented a single ribotype, whereas less-virulent isolates were genetically heterogeneous. Similarly Smith et al. (42) found that virulent isolates of serotypes 1 and 2 had distinct ribotype profiles, while Rasmussen et al. (37) demonstrated that particular ribotype profiles were clearly associated with clinical-pathology observations. A recent study of 99 strains by macrorestriction of DNA revealed four major clusters, one of which was strongly associated with invasive disease, and indicated that isolates from pigs with meningitis and septicemia showed a significantly higher degree of genetic homogeneity than isolates from pigs with pneumonia and healthy pigs (1).
Many basic questions about the population biology of S. suis and the nature, origin, and spread of virulent clones could be answered if isolates of this species could be characterized unambiguously. As S. suis is responsible for a range of diseases and can be carried asymptomatically, a study of the population structure of this organism could reveal clones or clonal groups with an apparently increased capacity to cause disease or an increased association with particular clinical manifestations of S. suis. As well as helping understand the epidemiology of S. suis infection and the biological relevance of the current serotyping approach, such work could facilitate the rapid identification of potentially virulent strains within herds, which could then be treated prophylactically. Multilocus sequence typing (MLST) is a highly discriminatory and unambiguous method of characterizing bacterial isolates that has now been successfully employed in the characterization of several species (10, 11, 12, 13, 29). MLST is based on the nucleotide sequences of internal fragments of housekeeping genes, in which mutations are assumed to be largely neutral (40). For each gene fragment the different nucleotide sequences are assigned allele numbers and the sequence type (ST) of each isolate is defined by the alleles present at each of seven distinct loci. Isolates that share the same ST are assumed to be members of the same clone, that is, they have a recent common ancestor. Due to the high numbers of alleles at each of the seven loci it is highly unlikely that isolates will have the same profile by chance. An important advantage of MLST is that sequence data are portable and can be readily compared among laboratories. In addition, the data obtained can be used to address questions about the evolutionary and population biology of bacterial species (14, 45).
Here we describe the development of an MLST scheme for S. suis. The scheme is based on the nucleotide sequences of seven housekeeping gene fragments and was used to evaluate 294 isolates from diverse geographical backgrounds. The isolates were derived from both asymptomatic carriage and from a range of disease states and represent some 28 different serotypes. The relationship of ST with serotype, currently the standard epidemiological marker for S. suis, is described, and additionally significant differences in the distributions of strains isolated from different disease states among clonal complexes are discussed.
MATERIALS AND METHODS
Bacterial isolates.
A total of 301 S. suis isolates were used in this study. Reference strains of serotypes 1 and 2 through 34 were supplied by L. A. Devriese (Faculty of Veterinary Medicine, University of Ghent, Ghent, Belgium), M. Gottschalk (Faculté de Médicine Vétérinaire, Université de Montréal, Montreal, Canada), and P. Heath (Veterinary Laboratories Agency, Bury Saint Edmunds, United Kingdom) (7). Twenty-one well-characterized serotype 2 isolates, including many isolates characterized in previous virulence studies, and 11 serotype 14 isolates from cases of meningitis were supplied by P. Norton (Institute for Animal Health, Newbury, United Kingdom). One hundred thirty-six field isolates, obtained by the Veterinary Laboratories Agency from diverse geographical sources across England, were included in this study. These were selected to represent isolates from diverse serotypes, sites of isolation, and clinical backgrounds, including invasive-disease isolates (meningitis, septicemia, and arthritis) and lung isolates from cases of pneumonia or other respiratory problems. One further isolate from the Warwick strain collection obtained from a case of porcine septicemia in the United Kingdom was included. A similar sample of 81 Spanish field isolates obtained by C. Tarradas and I. Luque was included in this study. The sample consisted of 38 “carried” isolates from the tonsils of healthy pigs and 43 clinical isolates, again with diverse clinical backgrounds (27). Two porcine isolates, one previously described as atypical (26), provided by C. Lammler, Institut für Tierarztliche, Giessen, Germany, were included. Fifteen isolates obtained from S. suis disease of humans were included in addition to the serotype 14 type strain, which was also originally a human isolate. These isolates were obtained from Augustine Cheng (Department of Microbiology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong; six isolates), M. Gottschalk (three isolates), G. Grise (Centre Hosptialier d'Elbeuf, Elbeuf, France; three isolates), C. Lammler (one isolate), P. Heath (one isolate), and B. Francois, Hospital Universitaire Dupuytren, Limoges, France (one isolate; B. Francois, V. Gissot, M. C. Ploy, and P. Vignon, Letter, J. Clin. Microbiol. 36:2395, 1998).
Preparation of chromosomal DNA.
Chromosomal DNA was prepared from all isolates as described previously (52).
Identification of housekeeping loci used for MLST.
Seven housekeeping gene loci were used in this study. The sequences of cpn60, encoding a 60-kDa chaperonin (6), dpr, a putative peroxide resistance protein (32), and recA, which encodes homologous recombination factor (48), were obtained from sequence databases. The sequences of aroA (encoding 5-enolpyruvylshikimate 3-phosphate synthase) and thrA (encoding aspartokinase/homoserine dehydrogenase) were kindly provided by A. Allen (University of Cambridge, Cambridge, United Kingdom). Finally the sequences for gki (encoding glucose kinase) and mutS (encoding a DNA mismatch repair enzyme) were obtained by us following PCR amplification and sequencing using primers designed to amplify the corresponding genes from other streptococci.
Amplification and nucleotide sequence determination.
PCR was performed under standard conditions with 30 cycles of 95°C for 1 min, X°C for 1 min, and 72°C for 1 min per kilobase of predicted product size, where X°C represents an annealing temperature appropriate for the particular primer set used. Products were visualized by agarose gel electrophoresis on 1.0% agarose in the presence of 1 μg of ethidium bromide ml−1. Details of all oligonucleotides used in this study are given in Table 1. PCR products were purified by passage through QiaQuick PCR product purification columns (Qiagen) and directly sequenced from both ends with the Beckman CEQ2000 system according to the manufacturer's instructions. Sequences were analyzed with the DNASTAR software, and in-frame internal fragments of the genes were selected for use in the MLST scheme.
TABLE 1.
Primers used for amplification and sequencing of the seven loci included in the S. suis MLST scheme
| Locus | Sequence
|
|
|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |
| dpr | CGTCTTTCAGCCCGCGTCCA | GACCAAGTTCTGCCTGCAGC |
| thrAa | GATTCAGAACGTCGCTTTGT | AAGTTTTCATAGAGGTCAGC |
| cpn60 | TTGAAAAACGTRACKGCAGGTGC | ACGTTGAAIGTACCACGAATC |
| recA | TATGATGAGTCAGGCCATG | CGCTTAGCATTTTCAGAACC |
| gki | GGAGCCTATAACCTCAACTGG | AAGAACGATGTAGGCAGGATT |
| aroA | TTCCATGTGCTTGAGTCGCTA | ACGTGACCTACCTCCGTTGAC |
| mutS | CGCAGAGCAGATGGAAGATCC | CCCATAGCTGTTTTGGTTTCATC |
The primer 5′-AAGAATGGATCATCAACCGT-3′ was used for the forward thrA sequencing reaction.
Allele and ST assignment.
For each locus distinct allele sequences were assigned arbitrary allele numbers with no weighting given to the degree of sequence divergence among alleles. For each isolate, the alleles at each of the seven loci defined the allelic profile or ST. The STs were assigned arbitrary numbers in order of description. STs were grouped into lineages or clonal complexes with the program BURST written and developed by E. Feil and located in the START, version 1.05, package of programs developed by K. Jolley (http://outbreak.ceid.ox.ac.uk/software.htm). The members of a lineage were defined as groups of two or more independent isolates that shared identical alleles at five or more loci. Each lineage was named after the ST identified as a putative ancestral type by BURST followed by “complex.” If no ancestral type was identified, the lineage was named after the STs contained within the clonal complex, for example, the ST-2/55 complex.
Confirmation of serotype.
The vast majority of isolates had been serotyped prior to receipt. However, in cases where an ST contained multiple serotypes, the serotypes were confirmed, where possible, by a serotype-specific PCR assay developed by Smith et al. (43, 44), which allows identification of the most common serotypes, 1 (plus 14), 2 (plus 1/2), 7, and 9.
Computational analyses.
The degree of clonality within the data set was estimated by calculating the index of association (IA) and its significance for all STs and for a subset of STs representative of each clonal complex by using a program written by Keith Jolley (http://www.mlst.net/indexassoc/indexassoc.htm). The determination of the number of nucleotide polymorphic sites, the calculation of dN/dS (where dN represents nonsynonymous base substitutions and dS represents synonymous base substitutions), and the construction of the dendrogram by using UPGMA (unweighted pair group method with arithmetic mean) were performed by using START (http://outbreak.ceid.ox.ac.uk/software.htm). The number of amino acid alterations, the maximum percent nucleotide divergence, and the average percent nucleotide divergence among alleles at a given locus were calculated by using the MEGA package, version 1.0 (S. Kumar, K. Tamura, and M. Nei, MEGA—molecular evolutionary genetics analysis 1.01, Pennsylvania State University, University Park, Pa., 1993). The test of Sawyer (39) was applied to the synonymous polymorphic sites within the alleles at each locus, and the significance of any clustering of polymorphic sites was evaluated by using 10,000 resamplings of the data.
RESULTS
Development of an MLST scheme for S. suis.
Chromosomal DNA was obtained from the 301 isolates, and the seven housekeeping gene loci from 294 isolates including the reference strains of 27 serotypes (serotypes 1 to 19, 23 to 25, and 27 through 31) were amplified. One or more housekeeping genes from the type strains of seven serotypes (20 through 22, 26, and 32 through 34) could not be amplified, and thus these serotypes were not examined further in this study. For all the remaining 294 isolates the sequences of the seven loci were determined and allelic profiles were assigned. The alleles defined for the MLST scheme were based on sequences of between 318 (cpn60) and 354 (recA) nucleotides, and between 32 (thrA) and 46 (gki) alleles per locus were present. The proportion of variable nucleotide sites present in the selected housekeeping genes ranged from 11.0 (thrA) to 29.0% (gki) (Table 2), while the number of polymorphic amino acid sites ranged from 3.4 (recA) to 17.9% (dpr). The proportions of nucleotide alterations that changed the amino acid sequence (dN) and the proportions of silent changes (dS) were calculated for each gene. From these data the dN/dS ratios for all seven loci were calculated, and all were substantially less than 1 (Table 2). For the 294 S. suis isolates, the mean number of alleles per locus was 40, providing the theoretical potential to distinguish >1.6 × 1011 different genotypes.
TABLE 2.
Characteristics of housekeeping gene loci included in the S. suis MLST scheme
| Locus | Putative function of gene product | Size of sequenced fragment (bp) | No. of alleles identified | No. of polymorphic nucleotide sites (%) | No. of polymorphic amino acid sites (%) | Nucleotide divergence between pairs of alleles (%)
|
dN/dS | GenBank accession No. | |
|---|---|---|---|---|---|---|---|---|---|
| Maximum | Avg | ||||||||
| aroA | EPSP synthase | 366 | 39 | 54 (14.8) | 20 (16.4) | 16 (4.4) | 7.5 | 0.0812 | AJ491619-AJ491657 |
| cpn60 | 60-kDa chaperonin | 318 | 43 | 79 (24.8) | 8 (7.6) | 39 (12.2) | 21.1 | 0.0063 | AJ491576-AJ491618 |
| dpr | Peroxide resistance | 336 | 37 | 61 (18.2) | 20 (17.9) | 24 (7.1) | 9.6 | 0.0957 | AJ491539-AJ491575 |
| gki | Glucose kinase | 321 | 46 | 93 (29.0) | 9 (8.4) | 54 (16.8) | 20.4 | 0.0174 | AJ491493-AJ491538 |
| mutS | DNA mismatch repair enzyme | 339 | 42 | 89 (26.3) | 12 (10.6) | 41 (12.1) | 18.5 | 0.0153 | AJ491451-AJ491492 |
| recA | Homologous recombination factor | 354 | 41 | 53 (15.0) | 4 (3.4) | 32 (9.0) | 10.9 | 0.0147 | AJ491373-AJ491413 |
| thrA | Aspartokinase/homoserine dehydrogenase | 336 | 32 | 37 (11.0) | 11 (9.8) | 10 (3.0) | 4.4 | 0.0715 | AJ491341-AJ491372 |
Relatedness of S. suis isolates.
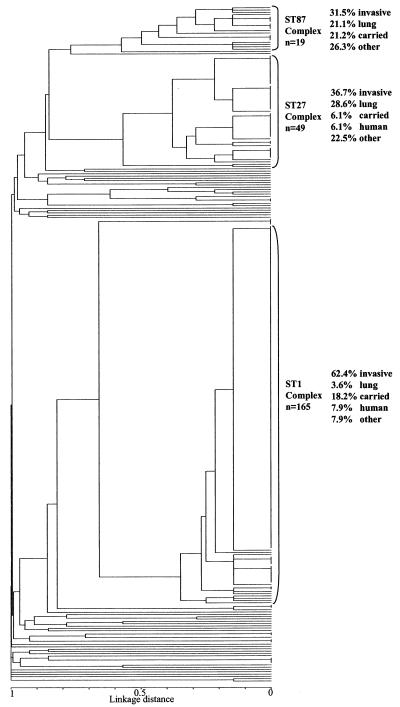
Figure 1 shows a dendrogram constructed from the matrix of pairwise allelic differences between the STs of all 294 isolates. The isolates resolved into 92 STs, 74 of which (80.4%) were identified only once. The most common ST, ST1, was identified 141 times in the data set, while 17 other STs contained between 2 and 13 members. Assignment of STs to lineages with BURST revealed that 37 were both unique and unrelated to any others, while the remaining 55 were assigned to 10 lineages (Table 3). complex was the largest and contained 165 isolates representing 14 STs. The ST27 complex included 49 members, the ST87 complex comprised 19 isolates, and the ST61 complex contained 4 isolates. The remaining six lineages contained two member STs (Table 3).
FIG. 1.
Dendrogram of 294 isolates of S. suis isolates constructed by UPMGA cluster analysis. Ninety-two STs were identified and three major clonal complexes were defined by using the program BURST. The members of a lineage were defined as groups of two or more independent isolates that had identical alleles at five or more loci. Percentages of isolates from the three major complexes isolated from invasive disease (meningitis, septicemia, and arthritis), pneumonia and other respiratory disease, porcine carriage, human disease, and other states (including reference strains previously used in virulence studies and isolates of unknown origin and those from other disease states) are indicated.
TABLE 3.
Characteristics of isolates belonging to the 92 STs identified in this study
| ST | ST profilea | No. | Serotype(s) (no.)b | Source (no.)c | Origind | ST complex |
|---|---|---|---|---|---|---|
| 1 | 1, 1, 1, 1, 1, 1, 1 | 141 | 1 (9), 8 (1), 1/14 (9), 14 (35), 2 (81)#, 1/2 (2), NK (4) | Invasive (94), carried (22), other (1), lung (5), human (10), NK (2), DP (1), RS (6) | HK, UK, USA, NL, SP, France | ST1 |
| 2 | 1, 1, 1, 2, 1, 1, 1 | 8 | 2 (8) | Invasive (5), carried (3) | UK | ST1 |
| 3 | 1, 1, 1, 16, 1, 1, 1 | 4 | 3 (4) | Carried (4) | SP | ST1 |
| 4 | 9, 1, 1, 1, 1, 1, 1 | 1 | NK (1) | Carried (1) | SP | ST1 |
| 5 | 1, 1, 1, 40, 1, 1, 1 | 1 | 2 (1) | Invasive (1) | SP | ST1 |
| 6 | 1, 1, 1, 1, 1, 1, 2 | 2 | 14 (2)# | Human (2) | Can, NL | ST1 |
| 7 | 1, 1, 1, 1, 1, 1, 3 | 1 | 2 (1) | Human (1) | HK | ST1 |
| 8 | 1, 1, 1, 1, 28, 1, 1 | 1 | 2 (1) | RS (1) | NL | ST1 |
| 9 | 1, 1, 1, 35, 1, 1, 1 | 1 | 2 (1) | Lung (1) | UK | ST1 |
| 10 | 1, 10, 1, 1, 1, 1, 1 | 1 | 1/14 (1) | Invasive (1) | UK | ST1 |
| 11 | 3, 1, 1, 1, 1, 1, 1 | 1 | 1/14 (1) | Invasive (1) | UK | ST1 |
| 12 | 3, 1, 1, 1, 1, 2, 1 | 1 | 1 (1) | Invasive (1) | UK | ST1 |
| 13 | 1, 12, 1, 1, 6, 21, 21 | 3 | 14 (1), 1 (2)# | Invasive (2), DP (1) | UK, Can, NL | Unrelated |
| 14 | 18, 1, 5, 12, 10, 1, 1 | 1 | 3 (1) | Lung (1) | SP | ST87 |
| 15 | 8, 8, 5, 12, 1, 10, 4 | 1 | 3 (1) | Invasive (1) | SP | ST87 |
| 16 | 5, 17, 5, 12, 1, 10, 4 | 3 | 4 (1), 9 (2) | Invasive (3) | SP | ST87 |
| 17 | 8, 1, 5, 12, 1, 10, 1 | 1 | 5 (1) | Invasive (1) | SP | ST87 |
| 18 | 31, 1, 5, 12, 1, 10, 1 | 1 | 8 (1) | Carried (1) | SP | ST87 |
| 19 | 1, 8, 24, 12, 1, 10, 4 | 3 | 2 (3) | Carried (1), RS (2) | NL | ST87 |
| 20 | 1, 1, 5, 12, 1, 10, 1 | 1 | 2 (1) | Carried (1) | NL | ST87 |
| 21 | 18, 8, 24, 12, 1, 9, 4 | 1 | 28 (1) | Lung (1) | UK | ST87 |
| 22 | 18, 8, 24, 12, 1, 11, 4 | 1 | 8 (1) | NK (1) | UK | ST87 |
| 23 | 26, 8, 24, 12, 1, 10, 4 | 3 | 4 (3) | Lung (2), other (1) | UK | ST87 |
| 24 | 6, 21, 24, 12, 1, 10, 4 | 1 | 7 (1) | Carried (1) | SP | ST87 |
| 25 | 9, 30, 5, 34, 30, 3, 25 | 13 | 2 (13) | Human (3), invasive (6), lung (1) RS (3) | UK, Can | ST27 |
| 26 | 2, 26, 5, 34, 31, 3, 25 | 1 | 2 (1) | RS (1) | USA | ST27 |
| 27 | 31, 30, 5, 34, 31, 3, 25 | 5 | 2 (2), 3 (3) | Invasive (4), lung (1) | Fin, UK, SP | ST27 |
| 28 | 2, 30, 5, 34, 31, 3, 25 | 11 | 2 (6), 1/2 (3), NT (2) | NK (1), lung (2), carried (2), invasive (5), RS (1) | Can, UK, SP | ST27 |
| 29 | 8, 30, 5, 34, 30, 3, 25 | 11 | 2 (2), 7 (7),# NT (1), 3 (1) | Lung (5), invasive (2), other (1), DP (1), carried (1), NK (1) | Fin, UK, Den | ST27 |
| 30 | 30, 30, 5, 34, 31, 3, 22 | 1 | NK (1) | Lung (1) | UK | ST27 |
| 31 | 30, 30, 5, 34, 31, 3, 25 | 2 | 3 (2) | Lung (2) | UK | ST27 |
| 32 | 8, 30, 5, 34, 30, 21, 4 | 1 | 7 (1) | Other (1) | UK | ST27 |
| 33 | 31, 30, 4, 34, 31, 3, 25 | 1 | 3 (1) | Lung (1) | UK | ST27 |
| 34 | 8, 43, 5, 34, 30, 21, 4 | 1 | 7 (1) | Lung (1) | UK | ST27 |
| 35 | 31, 30, 5, 34, 31, 4, 25 | 1 | 3# (1) | DP (1) | NL | ST27 |
| 36 | 24, 30, 35, 25, 9, 33, 20 | 1 | 3 (1) | Carried (1) | Fin | Unrelated |
| 37 | 25, 39, 5, 6, 14, 29, 12 | 1 | 2 (1) | Lung (1) | Fin | Unrelated |
| 38 | 31, 7, 8, 13, 6, 7, 24 | 1 | NK (1) | Invasive (1) | Fin | Unrelated |
| 39 | 21, 25, 19, 14, 5, 10, 9 | 2 | 5 (2) | Invasive (2) | UK | Unrelated |
| 40 | 37, 10, 14, 14, 8, 32, 27 | 1 | NT (1) | Other (1) | UK | Unrelated |
| 41 | 15, 27, 9, 4, 16, 26, 11 | 1 | 16 (1) | NK (1) | UK | Unrelated |
| 42 | 13, 13, 32, 28, 25, 38, 12 | 1 | 3 (1) | Lung (1) | UK | Unrelated |
| 43 | 35, 36, 27, 9, 37, 28, 29 | 1 | 15 (1) | Invasive (1) | UK | ST43/52 |
| 44 | 32, 19, 25, 10, 6, 12, 6 | 1 | 5 (1) | NK (1) | UK | Unrelated |
| 45 | 28, 31, 5, 7, 36, 24, 31 | 1 | 15 (1) | Invasive (1) | UK | Unrelated |
| 46 | 17, 3, 21, 7, 14, 6, 8 | 1 | 9 (1) | Other (1) | UK | ST46/50 |
| 47 | 25, 24, 9, 29, 7, 30, 12 | 1 | 16 (1) | Other (1) | UK | Unrelated |
| 48 | 37, 41, 20, 46, 38, 5, 27 | 2 | 9 (2) | Invasive (2) | UK | Unrelated |
| 49 | 31, 3, 16, 26, 35, 21, 6 | 1 | 15 (1) | Other (1) | UK | ST49 / 88 |
| 50 | 17, 42, 21, 15, 14, 6, 8 | 1 | 9 (1) | Invasive (1) | UK | ST46 / 50 |
| 51 | 29, 19, 26, 13, 6, 12, 20 | 1 | NK (1) | Other (1) | UK | Unrelated |
| 52 | 35, 36, 27, 9, 37, 18, 29 | 1 | 15 (1) | Invasive (1) | UK | ST43 / 52 |
| 53 | 8, 6, 18, 20, 28, 7, 5 | 1 | 5# (1) | DP (1) | Den | ST53 / 54 |
| 54 | 8, 6, 18, 20, 28, 7, 4 | 1 | 4# (1) | DP (1) | Den | ST53 / 54 |
| 55 | 25, 37, 15, 41, 15, 23, 10 | 1 | 6# (1) | DP (1) | Den | Unrelated |
| 56 | 1, 2, 17, 36, 29, 21, 15 | 2 | 1/2 (1), 2 (1) | Carried (2) | SP | ST56 / 58 |
| 57 | 27, 22, 11, 43, 12, 25, 18 | 1 | 10 (1) | Carried (1) | SP | Unrelated |
| 58 | 1, 2, 17, 37, 21, 15, 29 | 1 | 1 (1) | Carried (1) | SP | ST56 / 58 |
| 59 | 17, 21, 5, 44, 2, 22, 4 | 1 | 9 (1) | Invasive (1) | SP | ST61 |
| 60 | 31, 30, 18, 38, 17, 8, 15 | 1 | 15 (1) | Carried (1) | SP | Unrelated |
| 61 | 17, 21, 5, 45, 2, 22, 4 | 1 | 9 (1) | Carried (1) | SP | ST61 |
| 62 | 7, 31, 28, 8, 18, 16, 32 | 1 | 21 (1) | Carried (1) | SP | Unrelated |
| 63 | 19, 29, 12, 39, 24, 39, 16 | 1 | 15 (1) | Carried (1) | SP | Unrelated |
| 64 | 8, 21, 5, 45, 2, 22, 4 | 1 | 1/2 (1) | Invasive (1) | SP | ST61/PICK> |
| 65 | 10, 16, 23, 27, 23, 31, 17 | 3 | 15 (2), 27 (1) | Lung (3) | SP | Unrelated |
| 66 | 38, 35, 10, 42, 33, 17, 30 | 1 | 28 (1) | Carried (1) | SP | Unrelated |
| 67 | 27, 15, 11, 22, 12, 27, 11 | 1 | 10 (1) | Carried (1) | SP | Unrelated |
| 68 | 14, 20, 31, 27, 27, 34, 18 | 1 | 24# (1) | DP (1) | Can | Unrelated |
| 69 | 23, 33, 22, 30, 3, 15, 12 | 1 | 25# (1) | DP (1) | Can | Unrelated |
| 70 | 38, 28, 30, 15, 32, 26, 7 | 1 | 31# (1) | Other (1) | Can | Unrelated |
| 71 | 11, 5, 13, 11, 26, 36, 14 | 1 | 13# (1) | DP (1) | Den | Unrelated |
| 72 | 12, 9, 33, 3, 19, 35, 19 | 1 | 27# (1) | DP (1) | Can | Unrelated |
| 73 | 33, 37, 2, 19, 13, 5, 12 | 1 | 16# (1) | DP (1) | Den | Unrelated |
| 74 | 31, 2, 6, 31, 10, 14, 6 | 1 | 12# (1) | DP (1) | Den | Unrelated |
| 75 | 25, 32, 9, 15, 9, 13, 8 | 1 | 28# (1) | DP (1) | Can | Unrelated |
| 76 | 8, 23, 36, 24, 21, 22, 12 | 2 | 19# (1), 17# (1) | Carried (2) | Can | ST76/79 |
| 77 | 39, 38, 7, 32, 11, 3, 8 | 1 | 30# (1) | DP (1) | Can | Unrelated |
| 78 | 23, 11, 5, 7, 22, 8, 20 | 1 | 10# (1) | DP (1) | Den | Unrelated |
| 79 | 8, 21, 36, 45, 21, 12, 22 | 1 | 18# (1) | Carried (1) | Can | ST76/79 |
| 80 | 8, 21, 3, 45, 2, 22, 23 | 1 | 23# (1) | DP (1) | Can | ST61 |
| 81 | 22, 1, 20, 21, 4, 21, 26 | 1 | 15# (1) | DP (1) | NL | Unrelated |
| 82 | 25, 34, 29, 5, 34, 19, 28 | 1 | 9# (1) | DP (1) | Den | Unrelated |
| 83 | 8, 30, 5, 34, 39, 3, 25 | 1 | 7 (1) | Invasive (1) | UK | ST27 |
| 84 | 4, 1, 1, 1, 1, 1, 1 | 1 | NK (1) | NK (1) | Unknown | ST1 |
| 85 | 34, 14, 37, 33, 20, 40, 21 | 1 | NK (1) | NK (1) | Unknown | Unrelated |
| 86 | 1, 1, 1, 1, 1, 1, 25 | 1 | 2 (1) | NK (1) | SP | ST1 |
| 87 | 18, 8, 24, 12, 1, 10, 4 | 1 | 8# (1) | DP (1) | Den | ST87 |
| 88 | 36, 3, 16, 18, 35, 21, 6 | 1 | 11 (1) | DP (1) | UK | ST49/88 |
| 89 | 8, 8, 24, 12, 1, 10, 4 | 1 | 3 (1) | Invasive (1) | SP | ST87 |
| 90 | 20, 40, 5, 23, 40, 41, 1 | 1 | NK (1) | Invasive (1) | UK | Unrelated |
| 91 | 31, 18, 5, 12, 41, 10, 18 | 1 | 11# (1) | DP (1) | Den | Unrelated |
| 92 | 16, 4, 34, 17, 42, 37, 13 | 1 | 29# (1) | DP (1) | Can | Unrelated |
Allele numbers for each gene, presented in the following order: aroA, cpn60, dpr, gki, mutS, recA, thrA.
#, reference strain of a particular serotype. (7), represented by this ST. NK, not known; NT, not typeable.
Invasive isolates were obtained from cases of septicemia, meningitis, or arthritis. Lung isolates were obtained from cases of bronchopneumonia or other uncomplicated respiratory disease. NK, not known; DP, isolated from a diseased pig with no further details known; RS, reference isolate (14, which have been characterized previously in porcine models of infection, were included; see Table 4).
SP, Spain; HK, Hong Kong; UK, United Kingdom; Can, Canada; NL, The Netherlands; Fin, Finland; Den, Denmark; USA, United States.
Evidence of recombination.
The extent of recombination within the S. suis population was assessed by determining the IA (30). The IA for the complete data set was 4.874, but a value of 2.09 was obtained on reduction of the data set to a single representative of each ST. When randomized data sets (1,000 trials) were used, the latter value was significantly greater than zero, which would be the expected value for a population at linkage equilibrium (i.e., freely recombining). However, in populations in which recombination is sufficient to randomize alleles at different loci over a long time period, the recent expansion of clones can result in the appearance of multiple isolates with similar genotypes. Such a sampling bias is a particular problem in a population such as that described here, which is likely to be dominated by virulent clones which have risen to high frequency. To address this issue, the IA was recalculated by using one member of each lineage (47 STs). For this sample the IA was reduced to 0.230 and no significant linkage disequilibrium among alleles was observed. In support of the suggestion of the importance of recombination in the long-term evolution of S. suis, there was evidence of recombination within the cpn60, gki, and mutS genes, with the test of Sawyer (39) showing a highly nonrandom distribution of synonymous polymorphic sites (P ⩽ 0.0001 [sum of the squares of the condensed fragment lengths statistic]). There was no significant clustering of polymorphisms within the other gene fragments.
Relationship between STs and serotypes.
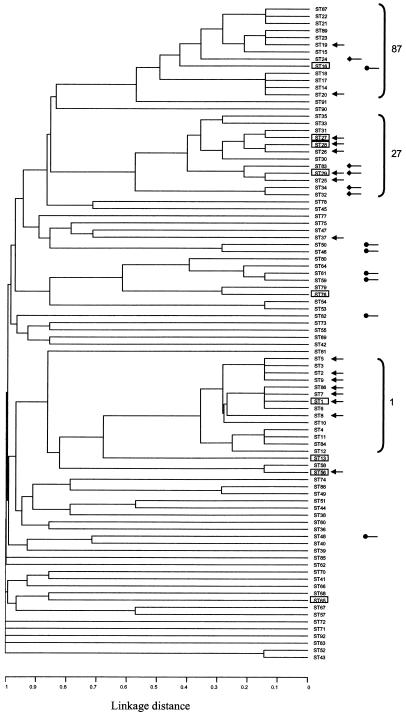
Of the 18 STs that contain more than one isolate 9 contained multiple serotypes (Table 3); the dendrogram (Fig. 2), which is reduced to showing only one member of each ST, highlights these 9 STs. When possible the serotypes of selected isolates were confirmed by PCR using specific primers (43, 44). Where large numbers of strains representing each different serotype were present in an individual ST (and thus there was little chance of the observed distribution reflecting serotyping errors), three representatives were selected for confirmatory PCR. Three of these STs contain serotypes which may, on the basis of the occurrence of some cross-reactivity in diagnostic serotyping, be closely related such as 2 and 1/2 or 1 and 14 (17). Thus these potential serotype exchanges should be treated with caution until more is known about the genetic relationships between the capsular loci of these serotypes. However, the remaining six STs contain serotypes with unambiguous serological responses which are not thought to be closely related to any other serotype.
FIG. 2.
Distribution of serotypes relative to ST. The dendrogram shown in Fig. 1 is reduced to show only one member of each ST. STs representing isolates of multiple serotypes are boxed (see Table 3). •, distribution of serotype 9; ♦, distribution of serotype 7; arrows, distribution of serotype 2. STs belonging to the three major clonal complexes are bracketed.
In the converse situation many serotypes were found to be associated with multiple STs. Thus, as illustrated in Fig. 2, serotype 2 isolates are represented in 16 of the 92 STs, serotype 9 isolates are represented in 7 distinct STs, and serotype 7 isolates are represented in 5 distinct STs. Again amplification with serotype-specific primers was used to confirm the wide distribution of serotypes 1 (plus 14), 2 (plus 1/2), 7, and 9. Hence, although many isolates of the same serotypes are closely related, these data clearly highlight that the serotype can be a poor marker of genetic relatedness of S. suis isolates.
Relationships among ST, disease state, and country of isolation.
The most frequently isolated ST, ST1, was identified in six countries and hence appears to be globally distributed (Table 3). Of the other 17 STs that include multiple isolates 6 were also found to contain strains from more than one country. Isolates belonging to the five STs containing five or more isolates originated from a diversity of sources. Strains isolated from different clinical states are widely distributed throughout the dendrogram; however, there is some evidence that isolates from different clone complexes have differing propensities to cause particular disease states. Thus an examination of Fig. 1 shows that the ST1 complex contains a substantially higher proportion of isolates from meningitis, septicemia, and arthritis than the ST27 and ST87 complexes. Chi-square tests comparing the numbers of invasive isolates with the numbers of other isolates in particular complexes indicate that the ST1 complex contains a significantly higher number of isolates from invasive disease than the ST87 (P < 0.01) and ST27 (P < 0.01) complexes. Both the ST87 (P < 0.01) and ST27 (P < 0.0001) complexes contain a significantly higher number of isolates from lung disease than the ST1 complex. The correlation between lung isolates and two individual STs appears even stronger. Five of the 11 ST29 isolates were isolated from the lung in cases of porcine pneumonia (an additional 2 of the 11 were also lung isolates, one from a case of septicemia and one from a sudden death, while the remaining 4 isolates include 1 from healthy carriage, 1 brain isolate, 1 from an unknown disease, and 1 unknown). Similarly all three ST65 isolates were from the lung. Carried isolates are widely distributed throughout the dendrogram and are represented in 22 of 92 STs. Sixteen isolates from human invasive disease were included in the study and were found to represent four STs, although the majority belong to ST1. Two of these four STs, ST1 and ST25, also contain a large number of isolates from various porcine disease states.
MLST findings in relation to isolates previously characterized in virulence studies.
Fourteen reference isolates included in this study had previously been included in studies of S. suis virulence using porcine infection models (Table 4) (33). The virulence of these strains had been determined by one of three different methodologies, either intravenously (i.v.) (33, 36) or by using the method of Vecht et al. (50) or the method of Galina et al. (15). The methods of both Vecht et al. (50) and Galina et al. (15) use intranasal inoculation of S. suis following inoculation with Bordetella bronchiseptica or porcine reproductive and respiratory syndrome virus, respectively. Six of the 14 isolates were representatives of ST1, and all 6 were defined as highly virulent in whichever model they were tested including both i.v. (3 isolates) and intranasal challenge (3 isolates). A further five isolates were members of the ST27 complex. Three ST25 isolates included isolates defined as of high, intermediate, or low virulence by i.v. inoculation, while two other members of the ST27 complex, an ST26 isolate and an ST28 isolate, were described as of low virulence in intranasal and i.v. challenges, respectively. ST19, represented by two isolates and a member of the ST87 complex, was associated only with strains of intermediate and low virulence as determined by intranasal challenge. Thus correlation of ST and observed virulence illustrates a trend suggesting that ST1 isolates are highly virulent and that isolates of other common STs, such as ST19 and ST25, may be somewhat less capable of causing invasive disease.
TABLE 4.
MLST analysis of isolates previously examined in porcine models of infection
| Strain | Virulence | ST (ST complex) |
|---|---|---|
| H11/1 | Highc | 1 (1) |
| D282 | Higha | 1 (1) |
| P1/7 | Highc | 1 (1) |
| 3881 | Higha | 1 (1) |
| 87555 | Highb | 1 (1) |
| B831 | Highc | 1 (1) |
| 1591 | Highd | 25 (27) |
| B554 | Intermediatec | 25 (27) |
| DH5 | Lowb | 26 (27) |
| TD-10 | Lowc, d | 25 (27) |
| 0891 | Lowd | 28 (27) |
| 3921 | Intermediatea | 19 (87) |
| 3898 | Lowa | 19 (87) |
| 3912 | Lowa | 8 (1) |
DISCUSSION
The primary objective of this work was to increase understanding of the population structure of S. suis by developing an unambiguous typing scheme which could then be expanded by other investigators and in turn to use this as a framework to help understand the differential virulence of S. suis isolates. Such a framework can be used as a basis on which the distribution and alleles of virulence genes of S. suis can be superimposed. Some of these genes, such as the suilysin gene, are already known to be absent from many S. suis isolates (25), and studies may identify genes (or alleles of genes) of particular importance in S. suis pathogenesis, which will in turn aid studies aimed at developing more-effective vaccines and therapeutics against S. suis. Previous studies of S. suis genetic diversity have largely included strains of only one serotype or isolates from a single country and have generally applied typing methods of low resolution that pose problems of reproducibility among laboratories. This study, however, uses two large collections of strains from the United Kingdom and Spain and includes smaller numbers of isolates from seven other countries. In addition this study utilizes MLST, which affords high discrimination, reproducibility, and potential accessibility of data over the Internet. MLST has already been used successfully in the characterization of other bacteria and has been validated against other molecular typing methodologies (10, 11, 12, 13, 29).
To facilitate this study, seven loci that could be amplified and sequenced from a wide range of S. suis isolates were chosen. The seven loci were not subject to positive selection, as demonstrated by the dN/dS ratio for each locus, which was substantially less than 1 (a dN/dS ratio >1 implies selection for amino acid change). For the 294 strains included in this study the number of alleles identified per locus was on average 40, which is consistent with previous phenotypic and genotypic analyses suggesting that S. suis represents a diverse species. The difficulty in amplifying all seven loci from some of the type strains is likely a reflection of this diversity. Type strains of seven serotypes (20 to 22, 26, and 32 to 34) were excluded from the study due to unsuccessful amplification of at least one housekeeping gene in each case. Six of these seven type strains have previously been shown to be more distantly related serotypes on the basis of 16S rDNA and cpn60 sequencing (6, 7). This study supports the argument that at least some of these serotypes may represent separate species (6). PCR amplification from the remaining type strain (serotype 21) was unsuccessful for a single locus, presumably due to divergence within this allele The IA, a measure of the extent of recombination for the complete data set, was 4.874, but this was reduced to 0.230 if one representative of each lineage was included. This value was not significantly different from the IA of zero expected for a population at linkage equilibrium. This implies that, although the observed population structure is clearly biased by the repeated recovery of recently arisen highly successful clones, in the longer term there is little clonal framework within the S. suis population. In support of this there was evidence of a history of horizontal gene transfer identified in cpn60, mutS, and gki by Sawyer's test (39).
In spite of the observed genetic diversity three major clonal groups dominate the S. suis population examined in this study. Most striking was the ST1 complex, containing 165 isolates, within which 141 isolates were found to represent ST1 itself. Isolates belonging to ST1 originated from six countries including several European nations, the United States, and Hong Kong, and a high proportion were associated with the classic S. suis invasive diseases, septicemia, meningitis, and arthritis. Such a high occurrence of one ST could suggest that the MLST scheme described here does not have a high power of discrimination. However, the mean number of alleles identified per locus was 40, providing the potential to distinguish >1.6 × 1011 different genotypes. Hence a more likely explanation is that ST1 isolates represent a highly successful clone which arose relatively recently and which has rapidly spread worldwide. Repeated recovery of such indistinguishable isolates from invasive disease in different countries clearly implies that STs, such as ST1, define strains with an increased capacity to cause disease. This might reflect a variety of factors such as increased fitness, the possession of certain virulence factors or allelic variants thereof, and the selection of clones possessing particular antibiotic resistance profiles by therapeutic or prophylactic use. Two other major complexes were apparent, the ST27 and ST87 complexes, although no individual ST was as dominant as ST1 within these complexes. Although isolates from septicemia and meningitis fall within these complexes, there appears to be a significantly higher proportion of lung isolates in the ST27 and ST87 complexes than in the ST1 complex. This observation could suggest that some S. suis strains may have a propensity to cause pneumonia and others, such as members of ST1, may be better equipped to cause the classic S. suis invasive diseases, septicemia, meningitis, and arthritis. However, as S. suis is often isolated from the lungs of pigs with pneumonia along with other agents potentially involved in respiratory disease (such as Pasteurella multocida, Haemophilus parasuis, Actinobacillus pleuropneumoniae, swine influenza virus, B. bronchiseptica, and Mycoplasma spp. [22, 28, 38]), the role of S. suis in pneumonia remains somewhat controversial. Although S. suis is isolated in pure culture from pneumonia with reasonable frequency (1, 22) further epidemiological studies are required to address the etiological role of S. suis in porcine pneumonia and the particular clones involved.
To help understand the relationships between clonal groups and virulence, 14 S. suis isolates which had previously been included in virulence studies using porcine models were included in this study (Table 4). Six of these isolates were found to belong to ST1, and, in agreement with the suggestion above that ST1 isolates are of high virulence potential, all were highly virulent in the models irrespective of whether infection was via the intranasal or the i.v. route. Five of the isolates were found to belong to the ST27 complex, members of which, on the basis of the epidemiological observations already discussed, may have a lower potential to cause classic invasive diseases such as septicemia and meningitis than members of the ST1 complex. The five ST27 complex isolates have been reported to have various virulence potentials (Table 4), with three members classified as of low virulence while two others were classified as of intermediate or high virulence. However the two studies that produced these results used i.v. inoculation, which bypasses the normal route of colonization and invasion via the nasopharyngeal membranes and the palatine tonsils (8, 53) and thus may not be an accurate indication of the potential to cause disease in the field. Two other isolates belonged to ST19, a member of the ST87 complex that again appears to contain isolates less strongly associated with invasive disease and that were described as being of low or intermediate virulence in an intranasal model of infection. Note that several discrepancies in the literature with regard to the virulence of individual isolates have previously been identified and that there have been conflicting findings in experimental infection models. Thus, for example, strain DH5 has been variously reported to be of high or low virulence (for a discussion see reference 25 and the letter from M. Gottschalk, R. Higgins, and S. Quessy [J. Clin. Microbiol. 37:4202-4203, 1999]). Conflicting data may reflect the age and immunological status of the animals used and/or the inoculum and the system used to score clinical symptoms and highlight the need for international agreement on a virulence model and virulent and avirulent control strains (17). However, in spite of these caveats, there appears to be some correlation between the observed virulence in experimental infection models and the observed distribution of disease-associated strains in this MLST study.
Previous studies using multilocus enzyme electrophoresis, DNA restriction endonuclease analysis, and ribotyping have demonstrated similarities between strains of S. suis from humans and pigs (3, 21, 31). In this study 16 human isolates were included; 10 belong to ST1, which, as already discussed, contains a large number of isolates from various porcine disease states. Of the remaining six, three are representatives of two STs containing only human isolates, which form part of the ST1 complex, while the remaining three belong to ST25, which is a member of the ST27 complex. The three human ST25 isolates were from Canada; this may reflect the prevailing clonal groups present there although further investigation of porcine isolates from Canada is required to confirm this. Similarly it is likely that the isolation of ST1 from humans, at least in Europe, reflects the prevalence of strains belonging to this ST in porcine invasive disease. There is therefore no convincing evidence for S. suis clones with an increased propensity to infect humans, and it seems that strains causing human infection may reflect the dominant clones in the local pig population.
Isolates of many of the most common serotypes were distributed widely across the dendrogram. Thus, for example, serotype 2 was found to be present in 16 of the 92 STs, and in the converse situation a number of STs which contain isolates of multiple serotypes were identified. Considerable genetic diversity between strains of the same serotype has previously been reported, and these observations make the use of serotyping as a means of strain identification for epidemiological studies unreliable. While many of these STs are closely related and therefore most likely descended from a recent common ancestor, the same serotype is seen in isolates which differ at all seven loci. One implication of these observations is that capsular genes, which specify serotype, may be spread horizontally through the population. Horizontal spread of capsular genes has been demonstrated in Streptococcus pneumoniae (9), where, as in S. suis, capsular genes which are conserved between serotypes flank variable serotype-specific loci encoding antigenic specificity (43) and recombination between the conserved regions flanking the serotype-specific genes results in serotype exchange. For S. pneumoniae the proposed mechanism involves natural transformation although, as S. suis is not known to be naturally transformable, the mechanism here remains unknown. However, similar horizontal movement of surface markers such as the M protein is known to occur in other non-naturally transformable streptococci such as Streptococcus pyogenes (51). Isolates of serotype 2 are the most widely distributed across the dendrogram. This may simply reflect the higher number of serotype 2 isolates included in the study. Alternatively, as any serotype exchange presumably occurs during the colonization of the host with multiple serotypes as demonstrated previously (5, 41) and as serotype 2 is among the most frequently isolated serotypes (1, 54), it may be that these strains simply come into contact with strains of other serotypes more frequently. It is interesting to speculate that the recent epidemiology of S. suis in the United Kingdom may reflect such a serotype exchange event. As mentioned previously serotype 14 became increasingly associated with the types of invasive disease normally associated with serotype 2 S. suis during the 1990s. Virtually all serotype 14 isolates characterized in this study belonged to ST1, which, as already described, contains a large number of virulent isolates, the majority of which belong to serotype 2. Thus it is possible that selective pressures imposed by increasing immunity to the dominant serotype 2 may have resulted in the emergence of a virulent variant which has retained the highly successful ST1 genotype but which has acquired a type 14 capsular locus.
In conclusion, we have devised the first unambiguous typing system for S. suis. Three major complexes were identified. The dominant ST1 complex represented 165 isolates and was strongly associated with invasive disease (septicemia, meningitis, and arthritis), while members of the smaller ST27 and ST87 complexes appear less strongly associated with these diseases but may be associated with respiratory disease. The wide distribution of serotypes throughout the dendrogram and the identification of nine STs containing multiple serotypes suggest that the capsular genes may be moving horizontally through the population. It is hoped that this MLST framework will now be expanded to include isolates from other countries. It will be of particular interest to examine North American isolates, as it is hypothesized that serotype 2 isolates there have virulence potentials and virulence factors different from those of European isolates (17). In addition future studies will aim to further the understanding of S. suis pathogenesis by examining the distribution and allelic diversity of potential virulence determinants of S. suis relative to the MLST framework.
Acknowledgments
We gratefully acknowledge all of the colleagues listed in Materials and Methods, who provided isolates for inclusion in this study and sequence data prior to publication.
This work was supported by a project grant from the BBSRC (grant reference 88/S11598), and A.M.W. was supported by a Wellcome Trust Research Fellowship in Biodiversity (grant reference 053589).
REFERENCES
- 1.Allgaier, A., R. Goethe, H. J. Wisselink, H. Smith, and P. Valentin-Weigand. 2001. Relatedness of Streptococcus suis of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arends, J. P., and H. C. Zanen. 1988. Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10:131-137. [DOI] [PubMed] [Google Scholar]
- 3.Beaudoin, M., J. Harel, R. Higgins, M. Gottschalk, M. Frenette, and J. I. MacInnes. 1992. Molecular analysis of isolates of Streptococcus suis capsular type 2 by restriction-endonuclease-digested DNA separated on SDS-PAGE and by hybridisation with an rDNA probe. J. Gen. Microbiol. 138:2639-2645. [DOI] [PubMed] [Google Scholar]
- 4.Berthelot-Herault, F., H. Morvan, A. Keribin, M. Gottschalk, and M. Kobisch. 2000. Production of muraminidase-released protein (MRP), extracellular factor (EF) and suilysin by field isolates of Streptococcus suis capsular types 2, 1/2, 9,7 and 3 isolated from swine in France. Vet. Res. 31:473-479. [DOI] [PubMed] [Google Scholar]
- 5.Brisebois, L. M., R. Charlebois, R. Higgins, and M. Nadeau. 1989. Prevalence of Streptococcus suis in four to eight week old clinically healthy piglets. Can. J. Vet. Res. 54:174-177. [PMC free article] [PubMed] [Google Scholar]
- 6.Brousseau, R., J. E. Hill, G. Prefontaine, S. Goh, J. Harel, and S. Hemmingsen. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Env. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatellier, S., M. Gottschalk, R. Higgins, R. Brousseau, and J. Harel. 1999. Relatedness of Streptococcus suis serotype 2 isolates from different geographical origins as evaluated by molecular fingerprinting. J. Clin. Microbiol. 37:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clifton-Hadley, F. A., and T. J. L. Alexander. 1980. The carrier site and carrier rate of Streptococcus suis type 2 in pigs. Vet. Rec. 107:40-41. [DOI] [PubMed] [Google Scholar]
- 9.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchange at the capsular polysaccharide biosynthetic locus leads to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil, E. J., M. C. J. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 15.Galina, L., C. Pijoan, M. Sitjar, W. T. Christianson, K. Rossow, and J. E. Collins. 1994. Interaction between Streptococcus suis serotype 2 and porcine reproductive and respiratory syndrome virus in specific-pathogen-free piglets. Vet. Rec. 134:60-64. [DOI] [PubMed] [Google Scholar]
- 16.Gogolewski, R. P., R. W. Cook, and C. J. O'Connell. 1990. Streptococcus suis serotypes associated with disease in weaned pigs. Aust. Vet. J. 67:202-204. [DOI] [PubMed] [Google Scholar]
- 17.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259-272. [DOI] [PubMed] [Google Scholar]
- 18.Gottschalk, M., R. Higgins, M. Jacques, K. R. Mittal, and J. Henrichsen. 1989. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 27:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottschalk, M., R. Higgins, M. Jacques, M. Beaudoin, and J. Henrichsen. 1991. Isolation and characterization of Streptococcus suis capsular types 9-22. J. Vet. Diagn. Investig. 3:60-65. [DOI] [PubMed] [Google Scholar]
- 20.Gottschalk, M., R. Higgins, M. Jacques, M. Beaudoin, and J. Henrichsen. 1991. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 29:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampson, D. J., D. J. Trott, I. L. Clarke, C. G. Mwaniki, and I. D. Robertson. 1993. Population structure of Australian isolates of Streptococcus suis. J. Clin. Microbiol. 31:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, P. J., and B. W. Hunt. 2001. Streptococcus suis serotypes 3 to 28 associated with disease in pigs. Vet. Rec. 148:207-208. [DOI] [PubMed] [Google Scholar]
- 23.Higgins, R., and M. Gottschalk. 2000. Distribution of the Streptococcus suis capsular types in 1999. Can. Vet. J. 41:414.. [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins, R., M. Gottschalk, M. Boudreau, A. Lebrun, and J. Henrichsen. 1995. Description of six new capsular types (29-34) of Streptococcus suis. J. Vet. Diagn. Investig. 7:405-406. [DOI] [PubMed] [Google Scholar]
- 25.King, S. J., P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2001. Distribution and genetic diversity of suilysin in Streptococcus suis isolated from different diseases of pigs and characterization of the genetic basis of suilysin absence. Infect. Immun. 69:7572-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lammler, C., and R. Weiss. 1997. Characterisation of an unusual Streptococcus suis isolated from an aborted fetus of a pig. Med. Sci. Res. 25:263-264. [Google Scholar]
- 27.Luque, I., C. Tarradas, R. Astorga, A. Perea, H. J. Wisselink, and U. Vecht. 1999. The presence of muramidase released protein and extracellular factor protein in various serotypes of Streptococcus suis isolated from diseased and healthy pigs in Spain. Res. Vet. Sci. 66:69-72. [DOI] [PubMed] [Google Scholar]
- 28.Macinnes, J. I., and R. Desrosiers. 1999. Agents of the “suis-ide diseases” of swine Actinobacillus suis, Haemophilus suis and Streptococcus suis. Can. J. Vet. Res. 63:83-89. [PMC free article] [PubMed] [Google Scholar]
- 29.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maynard-Smith, J., N. H. Smith, M. O'Rouke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogollon, J. D., C. Pijoan, M. P. Murtaugh, E. L. Kaplan, J. E. Collins, and P. P. Cleary. 1990. Characterization of prototype and clinically defined strains of Streptococcus suis by genomic fingerprinting. J. Clin. Microbiol. 28:2462-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niven, D. F., and A. Ekins. 2001. Iron content of Streptococcus suis and evidence for a dpr homologue. Can. J. Microbiol. 47:412-416. [DOI] [PubMed] [Google Scholar]
- 33.Norton, P. M., C. Rolph, P. N. Ward, R. W. Bentley, and J. A. Leigh. 1999. Epithelial invasion and cell lysis by virulent strains of Streptococcus suis is enhanced by the presence of suilysin. FEMS Immunol. Med. Microbiol. 26:25-35. [DOI] [PubMed] [Google Scholar]
- 34.Okwumabua, O., J. Staats, and M. M. Chengappa. 1995. Detection of heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping). J. Clin. Microbiol. 33:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perch, B., K. B. Pedersen, and J. Henrichsen. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J. Clin. Microbiol. 17:993-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quessy, S., J. D. Dubreuil, M. Caya, and R. Higgins. 1995. Discrimination of virulent and avirulent Streptococcus suis capsular type 2 isolates from different geographical origins. Infect. Immun. 63:1975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen, S. R., F. M. Aarestrup, N. E. Jensen, and S. E. Jorsal. 1999. Associations of Streptococcus suis serotype 2 ribotype profiles with clinical disease and antimicrobial resistance. J. Clin. Microbiol. 37:404-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reams, R. Y., D. D. Harrington, L. T. Glickman, H. L. Thacker, and T. L. Bowersock. 1996. Multiple serotypes and strains of Streptococcus suis in naturally infected swine herds. J. Vet. Diagn. Investig. 8:119-121. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 40.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sihvonen, L., D. N. Kurl, and J. Salmela. 1986. Infection with Streptococcus suis serotypes 1 and 2 in the same diseased pig. Acta Vet. Scand. 27:626-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, H. E., M. Rijnsburger, N. Stockhofe-Zurwieden, H. J. Wisselink, U. Vecht, and M. A. Smits. 1997. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J. Clin. Microbiol. 35:1049-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, H. E., L. van Bruijnsvoort, H. Buijs, H. J. Wisselink, and M. A. Smits. 1999. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol. Lett. 178:265-270. [DOI] [PubMed] [Google Scholar]
- 44.Smith, H. E., V. Veenbergen, J. van der Velde, M. Damman, H. J. Wisselink, and M. A. Smits. 1999. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37:3146-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spratt, B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr. Opin. Microbiol. 2:312-316. [DOI] [PubMed] [Google Scholar]
- 46.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 47.Staats, J. J., B. L. Plattner, J. Nietfeld, S. Dritz, and M. M. Chengappa. 1998. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J. Clin. Microbiol. 36:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takamatsu, D., M. Osaki, and T. Sekizaki. 2001. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45:101-113. [DOI] [PubMed] [Google Scholar]
- 49.Tarradas, C., C. Borge, A. Arenas, A. Maldonado, R. Astorga, A. Miranda, and I. Luque. 2001. Suilysin production by Streptococcus suis isolated from diseased and healthy carrier pigs in Spain. Vet. Rec. 148:183-184. [DOI] [PubMed] [Google Scholar]
- 50.Vecht, U., J. P. Arends, E. J. van der Molen, and L. A. van Leengoed. 1989. Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am. J. Vet. Res. 50:1037-1043. [PubMed] [Google Scholar]
- 51.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14:619-631. [DOI] [PubMed] [Google Scholar]
- 52.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, D. M., G. H. Lawson, and A. C. Rowland. 1973. Streptococcal infection in piglets: the palatine tonsils as portals of entry for Streptococcus suis. Res. Vet. Sci. 15:352-362. [PubMed] [Google Scholar]
- 54.Wisselink, H. J., H. E. Smith, N. Stockhofe-Zurwieden, K. Peperkamp, and U. Vecht. 2000. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet. Microbiol. 74:237-248. [DOI] [PubMed] [Google Scholar]