Abstract
Six isolates of Mycobacterium avium of genotype dnaJ+ IS901− IS1311+ IS1245+ and serotypes 6 (n = 1), 6/9, (n = 2), and 9 (n = 3) were obtained within a 5-month period from a human immunodeficiency virus-negative patient treated for tuberculosis. The isolates were identified with PvuII restriction fragment length polymorphism (RFLP) analysis as a single IS1311 RFLP type and six different IS1245 RFLP types. Six separate colonies/clones obtained by subculture from each of the six isolates were tested for MICs of a set of 10 drugs. This report documents the appearance of isolates that are resistant to antimycobacterial drugs as the duration of therapy increases. Because isolates recovered from the patient following longer duration of treatment were more likely to be resistant to more antimycobacterial drugs, we would conclude that there was selection for antimycobacterial drug-resistant isolates. Analyses of all 36 clones identified three IS1311 and 22 IS1245 types forming three clusters. Tests of 105 environmental samples collected in the home and the work place of the patient yielded 16 mycobacterial isolates, of which one M. avium from soil was of genotype dnaJ+ IS901+ IS1311+ IS1245+ and serotype 2, and the second M. avium from a vacuum cleaner was of genotype dnaJ+ IS901− IS1311+ IS1245+ and serotype 9. Overall analyses of the results did not reveal any relation between serotype, RFLP type, and drug susceptibility. Based on the course of the disease in the patient and different serotypes, IS1311 and IS1245 RFLP types of isolates of M. avium we suppose represent polyclonal infection.
Even in the beginning of the 21st century, human mycobacterial infections presented a serious health problem (16, 25, 48). In addition to isolates of the Mycobacterium tuberculosis complex, the causative agents of human lung diseases include nontuberculous mycobacteria, of which mostly isolates of the M. avium complex have been identified. Most of the diagnosed cases are opportunistic infections affecting immunocompromised patients due to different factors, including human immunodeficiency virus (HIV) infection (13). The treatment of infections, caused mostly by M. avium complex isolates, is rather difficult due to primary resistance of the agent against most of the commonly used drugs (1, 11, 16).
Human infections by M. avium complex isolates and the subsequent diseases result mostly from exposure of an immunocompromised patient to a high M. avium complex infection pressure. The rather high prevalence of M. avium complex infections in HIV-AIDS patients, reaching up to 65%, results from the frequent occurrence of M. avium complex isolates in various environmental components (water, soil, dust, dung, raw vegetables) in which the isolates can survive for long periods or even propagate (19, 20, 23). Their circulation in the environment occurs via inanimate materials (in particular water, dust, and soil) and various wild or captive animals. M. avium complex isolates have been isolated from wild birds (18), small vertebrates (14), invertebrates (15), and poikilotherms (30). As to farm animals, isolates of M. avium complex were most frequently detected from poultry and swine (12, 35, 37, 49).
Various biological methods, including bioassays in chicks, serological typing, probes (Accu-Probes, San Diego, Calif.), PCR, restriction fragment length polymorphism (RFLP) analysis, and further molecular biological techniques, have been used for more accurate identification and detailed differentiation of M. avium complex isolates (27, 31, 36, 39, 41, 42, 45, 53). All 28 serotypes of M. avium complex yield positive reactions with the Accu-Probe M. avium complex but differ in virulence, as demonstrated by bioassays in chicks. In terms of this characteristic, they were divided in the early 1970s into three groups: group 1, fully virulent (milliary tuberculosis of parenchymatous tissues); group 2, intermediate virulent (lesions at the inoculation site, parenchymas free of damage); and group 3, avirulent (39).
Group 1 (fully virulent for birds) includes serotypes 1 through 3 (39) and PCR types IS901+, IS1311+, and IS1245+ (27, 36, 41). Isolates of the three serotypes reacted with the M. avium complex and M. avium probes and failed to react with the M. intracellulare probe (45, 46). M. avium complex isolates showing these characteristics were most frequently isolated from birds and swine and only very seldom from environmental samples and humans (36).
Group 2 (partially virulent for birds) includes serotypes 4 through 6, 8 through 11, and 21. Isolates of this group reacted with the same probes as those of group 1 (M. avium complex positive, M. intracellulare negative), but were classified by PCR with the genotype IS901− IS1311+ IS1245+ (39, 41, 43, 45, 46). Isolates showing these characteristics were isolated most frequently from human, porcine, and environmental samples (3, 6, 33, 36, 41).
Group 3 (avirulent for birds) included isolates of the remaining serotypes 7, 12 through 20, and 22 through 28. The isolates reacted with the probes M. avium complex and M. intracellulare but not with the probe M. avium and were classified by PCR with the genotype IS901− IS1311− IS1245− (6, 17, 39, 41, 45, 46). Most of the isolates were obtained from environmental samples (20).
RFLP analyses of all 28 serotypes of M. avium complex isolates carried out with the three insertion sequences IS901, IS1311, and IS1245 also resulted in classification of the serotypes with the groups defined above. IS901 was detected in the genome of isolates of group 1 in more than four copies at various positions (27, 41). IS1245 was present in three copies only, and their positions, designated the avian RFLP type, were invariable (6, 17, 33, 41). IS1311, which showed a high sequence homology with IS1245, was present in the genome of group 1 isolates in a single copy in the conservative position (33). RFLP analyses of group 2 isolates showed the presence of IS1245 and IS1311 in more than three copies in various positions.
The objective of our study was to analyze isolates of M. avium complex obtained from a female patient suffering from a lung infection by serotyping, PCR, and RFLP over time, to determine the MICs of selected antimycobacterial drugs for the isolates, and to find environmental sources of the infection in the home and work place of the patient.
CASE REPORT
The patient was a 47-year-old HIV-negative female affected by a serious deficiency of cellular immunity and suffering from M. avium complex-induced lung mycobacteriosis. The initial treatment with antimycobacterial drugs (isonikotine hydrazide, rifampin, pyrazinamide, and ethambutol) resulted in only partial mitigation of clinical manifestations; clearance of mycobacteria and complete retreat of clinical disease was achieved only when drug therapy was completed by immunostimulation. The clinical course and therapy with other antimycobacterial drugs (streptomycin, ciprofloxacin, clarithromycin, rifabutin, and amikacin) are described in detail by Ostadal et al. (34).
The patient showed a decrease in the number of CD4+ lymphocytes to 0.29 × 109/liter, resulting in a marked imbalance of the CD4/8 lymphocytes, and a weak reaction to phytohemagglutinin in the lymphocyte transformation test. The patient was living in a village and working in the local agricultural cooperative as a cattlewoman. From the beginning of 1995 she was exposed to excessive physical and psychological strain and was suffering from lack of sleep. Medical care was sought in September 1995. The patient complained about an irritating cough with occasional mucopurulent expectoration, perspiration, subfebrile body temperature in the evening, loss of appetite, and fatigue.
MATERIALS AND METHODS
Mycobacterial examination. (i) Samples of biological materials.
Culture of sputum samples collected during 5 months of the treatment period yielded six isolates of M. avium complex that were put to detailed analyses. At the end of the therapy of the patient, a total of 105 samples were collected and tested within epidemiological tracing of the source of infection in the work place (n = 28) and the home of the patient (n = 77).
(ii) Examination by culture.
Sputum samples were homogenized, decontaminated with sodium laurylsulfate, and inoculated onto two egg media of Ogawa, one egg medium of Löwenstein-Jensen, and one liquid serum medium of Sula (26). Environmental samples were homogenized, decontaminated with HCl and NaOH, and inoculated onto one Herrold egg yolk medium, one egg medium of Stonebrink, and one liquid serum medium of Sula (15, 26, 52).
(iii) Identification of isolates.
The mycobacterial isolates from the sputum and environmental samples were identified by the Accu-Probe technique with the M. tuberculosis complex probe for species of the M. tuberculosis complex and the M. avium complex and M. intracellulare probes for species of the M. avium complex. The M. avium complex isolates and clones were also tested by serotyping (47, 53), biochemical tests (51), and molecular biological techniques PCR and RFLP, as described below. The homogeneity of seven M. avium complex isolates (six from sputum and one from the dust-collecting bag of a vacuum cleaner) was tested by subculturing six single colonies derived from the original isolates. Each culture received from one single colony was designated in our paper as a clone.
PCR.
The isolates were determined to be mycobacteria by the detection of the genus-specific dnaJ gene with the primers 5′-GGG TGA CGC GAC ATG GCC CA-3′ and 5′-CGG GTT TCG TCG TAC TCC TT-3′ (32). IS901 was identified with the primers 5′-GCA ACG GTT GTT GCT TGA AA-3′ and 5′-TGA TAC GCC CGC AAT CGC GT-3′ (27), IS1245 with the primers 5′-GCC GCC GAA ACG ATC TAC-3′ and 5′-AGG TGG CGT CGA GGA AGA-3′ (17), and IS1311 with the primers 5′-TGC TGG ACG CAT TAC GCA ATG-3′ and 5′-CCG CAA CTC CAA ATC GCC AG-3′ (8).
RFLP.
Mycobacterial DNA was isolated as described by van Soolingen et al. (50). DNA was isolated from at least three bacteriological loops. The bacterial mass was suspended in 400 μl of 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The suspension was completed with 50 μl of lysozyme (10 mg/ml) and incubated at 37°C for 3 h. Then, 75 μl of a 10% sodium dodecyl sulfate-proteinase K mixture was added, the suspension was incubated at 65°C for another 10 min, and cetyltrimethylammonium bromide was added to complete cell wall, protein, and polysaccharide degradation.
DNA was purified with a chloroform-isoamyl alcohol mixture and precipitated with isopropanol. The precipitate was dissolved in 1× TE buffer. Approximately 5 μg of the purified mycobacterial DNA was digested with restriction endonuclease PvuII. The resulting DNA fragments were separated by agarose gel electrophoresis, exposed in a UV transilluminator, and transferred from the gel by vacuum blot onto a nylon membrane. DNA was fixed and hybridized with the respective labeled probe.
Preparation of constructs pMA12 and pCR-2.1-IS1311.
Plasmid pMA12 with the cloned IS1245-specific fragment was provided by Pieter Overduin (National Institute of Public Health and the Environment, Bilthoven, The Netherlands). The plasmid was selected from the KspI/BamHI M. avium subsp. paratuberculosis genomic library. Approximately 250 fg of plasmid DNA was used for PCR. The positive M. avium isolate 543/95 was used for the preparation of the IS1311 probe. The IS1311 sequence of this isolate was transcribed by PCR with the primers specified above (8). The 199-bp PCR product was cloned into the vector pCR2.1 (TA cloning kit; Invitrogen), and the resulting construct, pCR2.1-IS1311, was used at 250 fg of plasmid DNA in PCRs for the preparation of a probe with the primers of Collins et al. (8).
Probe preparation.
All the original cultures and clones derived from them were tested with a nonradioactive probe prepared by PCR amplification of plasmid pMA12 for the preparation of a probe identifying the element IS1311 and of the construct pCR2.1-IS1311 for the preparation of a probe identifying the element IS1245. The resulting 426-bp amplification product was obtained with IS1245-specific primers derived as described by Guerrero et al. (17). A 199-bp product was obtained with the IS1311-specific primers described by Collins et al. (8). A check of the PCR amplification products in agarose gels was followed by purification of the fragments of anticipated sizes with the QIAquick gel extraction kit (Qiagen, Berlin, Germany), which were subsequently used as the hybridization probes. The probes were labeled according to the manufacturer's instructions (ECL direct labeling kit; Amersham, London, United Kingdom).
Designation of RFLP types and subtypes.
DNA fingerprints were scanned with a charge-coupled device camera (UltraLum KS4000) and analyzed with the software Gel Compar (version 4.1; Applied Maths, Kortrijk, Belgium) by the unweighted pair group method using arithmetic averages (Dice coefficient) method. IS1311 RFLP type was marked according to the origin of the isolate, sputum or dust. IS1311 RFLP subtypes of the clones were marked with Roman numerals. IS1245 RFLP types of the isolates were marked with capitals. New RFLP type of the isolate was defined by a difference in at least five bands. Differences in IS1245 RFLP type between RFLP type A detected in the first isolate and the subsequent RFLP types are shown by arrows (Fig. 2). IS1245 RFLP subtype received from the clone was marked with the capital corresponing ot the RFLP type of the original isolate and an Arabic numeral expressing the heterogeneity.
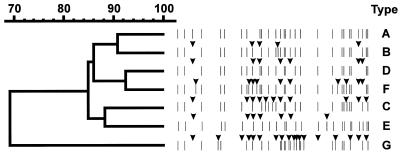
FIG. 2.
Dendrogram of IS1245 RFLP types of sputum isolates (RFLP types A to F) and the single dust isolate (RFLP type G). The arrows indicate differences in RFLP profile between RFLP type A detected in the first isolate and subsequent RFLP types.
Determination of MICs of selected antimycobacterial drugs.
The susceptibilities of the 36 clones derived from isolates from sputum to 10 drugs were determined by the quantitative micromethod MIC in Sula's medium (22). The MIC was defined as the lowest drug concentration causing visible inhibition of growth. Each susceptible pattern was designated by a lowercase letter.
RESULTS
Demonstration of mycobacteria in sputum.
None of the six mycobacterial isolates from the patient during a 5-month treatment period reacted with the M. tuberculosis complex or M. intracellulare Accu-Probe, and all the isolates reacted with the M. avium complex and probes as M. avium. The isolates and clones were identified as genotype dnaJ+ IS901− IS1311+ IS1245+. Isolates were of serotypes 6 (n = 1), 6/9 (n = 2), and 9 (n = 3), and clones were of the same serotypes: 6 (n = 3), 6/9 (n = 18), and 9 (n = 15) (Table 1).
TABLE 1.
MICs for all clones derived from sputum isolates
| Isolate serotype | IS-RFLP
|
Clone serotype | MICa (μg/ml)
|
Susceptibility patternb | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date of isolation (mo/day/yr) | Type | Subtype
|
Cluster (clade) | INH | RIF | ETB | STR | GEN | OFX | AMK | RFB | CLO | CLR | ||||
| 1245 | 1311 | ||||||||||||||||
| 6 | 11/20/95 | A | A1 | I | I (b) | 6/9 | 4 | 8 | 16 | 4 | 2 | 4 | 2 | 1 | 0.06 | 4 | a |
| A2 | III | III | 6 | 4 | 8 | 16 | 4 | 1 | 4 | 1 | 1 | 0.06 | 4 | a | |||
| A3 | I | I (a) | 6 | 8 | 8 | 16 | 4 | 1 | 4 | 1 | 2 | 0.06 | 2 | a | |||
| A3 | I | I (a) | 6 | 8 | 8 | 16 | 2 | 1 | 4 | 1 | 1 | 0.06 | 2 | b | |||
| A4 | I | I (a) | 6/9 | 4 | 8 | 16 | 2 | 1 | 4 | 1 | 1 | 0.03 | 2 | b | |||
| A4 | I | I (a) | 6/9 | 4 | 8 | 16 | 4 | 1 | 4 | 1 | 1 | 0.06 | 1 | a | |||
| 6/9 | 12/21/95 | B | A1 | I | I (b) | 6/9 | 16 | 1 | 16 | 2 | 1 | 4 | 1 | 0.50 | 0.06 | 1 | b |
| B1 | I | I (a) | 6/9 | 8 | 0.5 | 8 | 1 | 1 | 2 | 1 | 0.13 | 0.03 | 1 | c | |||
| B2 | I | I (a) | 6/9 | 8 | 0.5 | 8 | 1 | 1 | 2 | 1 | 0.13 | 0.06 | 2 | c | |||
| B2 | I | I (a) | 9 | 8 | 1 | 8 | 1 | 1 | 2 | 1 | 0.50 | 0.06 | 2 | d | |||
| B3 | I | I (a) | 9 | 8 | 0.5 | 8 | 2 | 0.5 | 2 | 1 | 0.13 | 0.06 | 1 | c | |||
| B4 | I | I (a) | 9 | 8 | 0.5 | 8 | 1 | 1 | 2 | 1 | 0.13 | 0.06 | 2 | c | |||
| 9 | 1/4/96 | C | C1 | I | I (a) | 9 | 4 | 8 | 4 | 2 | 0.5 | 4 | 1 | 2 | 0.06 | 1 | b |
| C2 | I | I (a) | 9 | 16 | 8 | 16 | 8 | 2 | 8 | 1 | 2 | 0.06 | 4 | a | |||
| C2 | I | I (a) | 6/9 | 16 | 8 | 16 | 4 | 2 | 4 | 2 | 2 | 0.06 | 4 | a | |||
| C2 | I | I (a) | 6/9 | 8 | 8 | 8 | 2 | 1 | 8 | 1 | 2 | 0.06 | 1 | b | |||
| C2 | I | I (a) | 6/9 | 16 | 2 | 16 | 2 | 1 | 2 | 0.5 | 0.25 | 0.06 | 1 | d | |||
| C3 | I | I (a) | 6/9 | 16 | 8 | 16 | 1 | 2 | 2 | 1 | 2 | 0.06 | 1 | d | |||
| 6/9 | 1/18/96 | D | D1 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 2 | 0.13 | 4 | a |
| D2 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 2 | 8 | 2 | 2 | 0.06 | 4 | a | |||
| D2 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 1 | 8 | 2 | 1 | 0.06 | 4 | a | |||
| D3 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 2 | 8 | 2 | 1 | 0.13 | 4 | a | |||
| D4 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 2 | 1 | 0.13 | 4 | a | |||
| D5 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 2 | 1 | 0.06 | 4 | a | |||
| 9 | 4/12/96 | E | B3 | I | I (a) | 9 | 16 | 8 | 16 | 16 | 1 | 4 | 2 | 1 | 0.06 | 1 | a |
| E1 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 2 | 0.06 | 4 | a | |||
| E2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 2 | 0.06 | 4 | a | |||
| E2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 4 | 1 | 2 | 0.13 | 2 | a | |||
| E3 | II | II | 6/9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 1 | 0.06 | 4 | a | |||
| E4 | II | II | 6/9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 1 | 0.06 | 4 | a | |||
| 9 | 4/18/96 | F | F1 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 0.5 | 8 | 1 | 1 | 0.06 | 4 | a |
| F1 | I | I (b) | 6/9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 0.5 | 0.13 | 4 | a | |||
| F2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 0.5 | 8 | 2 | 0.5 | 0.06 | 4 | a | |||
| F2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 0.5 | 0.13 | 4 | a | |||
| F2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 2 | 1 | 0.13 | 4 | a | |||
| F2 | I | I (b) | 9 | 16 | 8 | 16 | 16 | 1 | 8 | 1 | 1 | 0.13 | 4 | a | |||
Abbreviations: INH, isonikotine hydrazide (isoniazid) (sensitive, 0.25 μg/ml; resistant, 0.5 μg/ml); RIF, rifampin (sensitive, 0.5 μg/ml; resistant, 1.0 μg/ml); ETB, ethambutol (sensitive, 2.0 μg/ml; resistant, 4.0 μg/ml); STR, streptomycin (sensitive, 2.0 μg/ml; resistant, 4.0 μg/ml); GEN, gentamicin (sensitive, 2.0 μg/ml; resistant, 4.0 μg/ml); OFX, ofloxacin (sensitive, 2.0 μg/ml; resistant, 4.0 μg/ml); AMK, amikacin (sensitive, 4.0 μg/ml; resistant, 8.0 μg/ml); RFB, rifabutin (ansamycin) (sensitive, 0.13 μg/ml; resistant, 0.25 μg/ml); CLO, clofazimin (lamprén) (sensitive, 0.13 μg/ml; resistant, 0.25 μg/ml); CLR, clarithromycin (sensitive, 0.13 μg/ml; resistant, 0.25 μg/ml). Concentrations of clarithromycin for sensitivity/resistance assessment were not determined.
Susceptibility patterns derived from MICs of nine drugs (clarithromycin not included).
PvuII RFLP analysis.
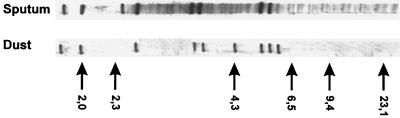
PvuII RFLP analysis, used for detailed differentiation of the isolates, yielded a single IS1311 RFLP type (Fig. 1) and six IS1245 RFLP types, marked A, B, C, D, E, and F (Fig. 2, Table 1). The subsequent RFLP analyses of six clones derived from one colony of each of the six isolates yielded three IS1311 RFLP subtypes, marked I, II, and III (Fig. 3, Table 1) and 22 IS1245 RFLP subtypes marked A1 through A4, B1 through B4, C1 through C3, D1 through D5, E1 through E4, F1, and F2 (Fig. 4, Table 1). The IS1245 RFLP analysis of clones of two B and E type isolates identified the RFLP type of one clone that had already been found in earlier isolates (Table 1).
FIG. 1.
IS1311 RFLP types of all sputum isolates and the single dust isolate. Sizes are shown in kilodaltons.
FIG. 3.
Dendrogram of IS1311 RFLP subtypes obtained by cloning of sputum and dust isolates.
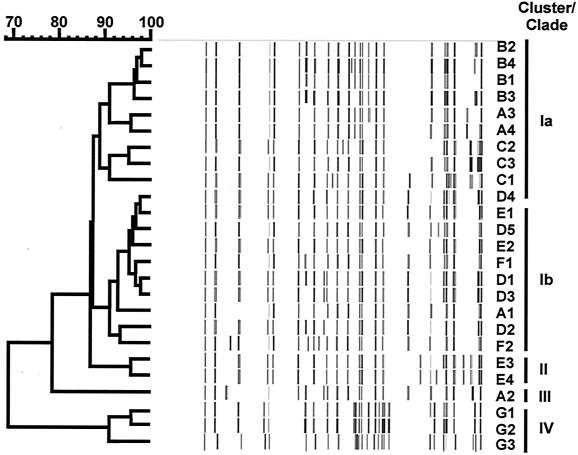
FIG. 4.
Dendrogram of IS1245 RFLP subtypes obtained by cloning of sputum and dust isolates.
Cluster analysis.
The IS1311 RFLP subtypes identified in the 36 clones could be grouped into three clusters (>79.5% matching patterns). The first cluster included 33 clones, the second included two, and the third included a single clone (Fig. 3, Table 1). Similarly, the IS1245 RFLP subtypes identified in the 36 clones could be grouped into three clusters (>69.3% matching patterns). The first cluster included 33 clones, the second cluster included two clones, and the third cluster included a single clone (Fig. 4, Table 1). Matching of the two dendrograms yielded clusters I, II, and III, of which cluster I could be subdivided into clades Ia and Ib (Fig. 4, Table 1).
Determination of MICs and evaluation of IS1311 and IS1245 RFLP analyses.
All 36 clones derived from the six isolates were resistant to isonikotine hydrazide and ethambutol (Table 1). Clones resistant to isonikotine hydrazide showed a progressive increase in MIC. The resistance of the clones to rifampin was stable during therapy except in the second month of therapy. The susceptibility of some clones to streptomycin was found only in isolates detected during the first 3 months of therapy. From the fourth month on, they became resistant. All the isolates were sensitive to gentamicin, amikacin, and clofazimin. Only some isolates received in the second and third months were sensitive to ofloxacin. In terms of susceptibility to nine drugs (clarithromycin not included), four susceptible patterns were defined, a, b, c, and d, which were found in 66.7%, 13.9%, 11.1%, and 11.1% of the clones, respectively (Table 1).
As for the IS1311 RFLP subtypes, the susceptible pattern a was found in 21 clones of subtype I, two clones of subtype II, and one clone of subtype III, and the patterns b, c, and d in 5, 4, and 3 clones of subtype I, respectively. As for IS1245, the susceptible pattern a was found in 18 clones at the beginning (A1 through A4, B3, and C2) and towards the end (D1 through D5, E1 through E4, F1, and F2) of the treatment period. The patterns b, c, and d were found in one clone each (C1, B1, and C3, respectively). The susceptible pattern of some IS1245 subtypes (A1, A3, A4, B2, B3, and C2) changed during the treatment period. Only pattern a was identified in clones of IS1311 RFLP subtypes II and III, as well as in IS1245 RFLP subtypes A2, E3, and E4, belonging to clusters II and III. On the other hand, all four susceptible patterns were found in the clones forming cluster I of the IS1245 RFLP subtypes.
Demonstration of mycobacteria in environmental samples.
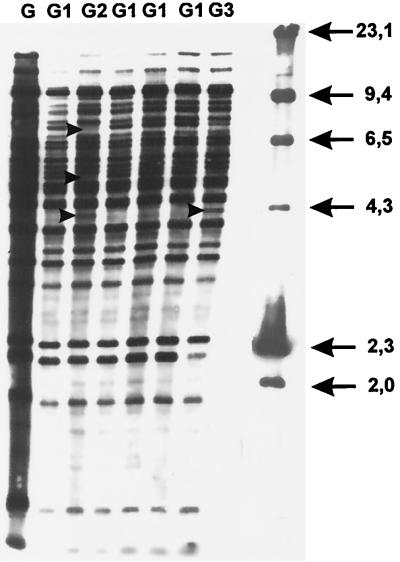
Examination of environmental samples yielded 16 dnaJ+ mycobacterial isolates, of which none reacted with the Accu-Probe M. tuberculosis complex and M. intracellulare probes and two reacted with the M. avium complex probe. Biochemical typing identified seven isolates as M. gordonae, two isolates as M. fortuitum, and two isolates as M. avium complex. Species of the remaining five dnaJ+ IS901− IS1311− IS1245− mycobacterial isolates were not identifiable (Table 2). The soil M. avium isolate was classified as serotype 2 and genotype dnaJ+ IS901+ IS1311+ IS1245+ and the dust isolate as serotype 9 and genotype dnaJ+ IS901− IS1311+ IS1245+ (Table 2). The IS1311 and IS1245 RFLP types of the dust isolate differed from those of the sputum isolates (Fig. 1 and 2). Six clones derived from the dust isolate were of serotype 9 and were classified with IS1311 as subtype IV and with IS1245 as subtypes G1 through G3 forming cluster IV (Fig. 3, 4, and 5).
TABLE 2.
Examination of environmental samples from the home and the work place
| Locality | Place | Biological material collected | No. of samples
|
Identification | |
|---|---|---|---|---|---|
| Examined | Positive | ||||
| Work place | Cattle house | Drinking water sediment | 6 | 5 | M. gordonae |
| Floor scrapings | 12 | 0 | |||
| Feed stuff | Silage | 2 | 0 | ||
| Courtyard | Invertebrates (fliesa) | 4 | 0 | ||
| Office | Drinking water sediment | 4 | 1 | M. gordonae | |
| Total (%) | 28 (100) | 6 (21.4) | |||
| Home | Bathroom | Water and shower biofilm | 9 | 1 | M. gordonae |
| 2 | M. fortuitum | ||||
| House corridor | Garden soil | 8 | 1 | M. avium complex (serotype 2)b | |
| Nursery | 3 | 1 | Mycobacterium sp.c | ||
| Living room | 3 | 1 | Mycobacterium sp. | ||
| Whole house | Dust from vacuum cleaner 1 month old | 30 | 1 | M. avium complex (serotype 9)d | |
| 1 | |||||
| Courtyard | Moss from the garden | 3 | 0 | Mycobacterium sp. | |
| Scraping under the eaves | 1 | 1 | Mycobacterium sp. | ||
| Earthworms | 1 | 0 | |||
| Barn | Floor dust | 17 | 1 | ||
| Poultry house | Hen's nest dust | 1 | 0 | Mycobacterium sp. | |
| Garden | Earthworms | 1 | 0 | ||
| Total (%) | 77 (100) | 10 (13.0) | |||
| Total (%) | 105 (100) | 16 (15.2) | |||
One sample with two imagoes of Muscidae and one sample with 126 imagoes of stable fly (Stomoxys calcitrans).
dnaJ+ IS901+ IS1311+ IS1245+.
Slowly growing nonpigmented dnaJ+ IS901− IS1311− IS1245− isolates.
dnaJ+ IS901− IS1311+ IS1245+.
DISCUSSION
The following three hypotheses of the source of the M. avium infection in our patient were considered: concurrent or consecutive infections by M. avium isolates belonging to different serotypes and genotypes which, in the case of repeated infections, may have come from the environment; reactivation of endogenous infection manifested by aggravation of clinical symptoms provided that characteristics of the isolates obtained at the beginning and towards the end of the infection would be identical; microevolution of isolates, which would be confirmed by progressive transformation of RFLP types. Genomic mutations might result from transposition of insertion elements or site-specific recombination between sequences of insertion elements or homologous sequences at insertion element hot spots.
The hypotheses were tested at the DNA level with the insertion sequences IS1311 and IS1245. The latter is a suitable marker for epidemiological studies because most human isolates, classified as serotypes 4, 6, and 8, yield polymorphic RFLP profiles with 6 to 20 bands. These RFLP types are designated nonavian (41). Knowledge of the in vivo and in vitro stability of the insertion elements is essential for interpretation of the results of epidemiological studies. In vitro stability of IS1245 has been demonstrated by serial passage in culture media (2, 38).
Absolute IS1245 stability has not been demonstrated (2, 38). Bauer et al. (3), who in 1999 compared the RFLP profiles obtained from colonies of several isolates obtained from a single HIV-positive patient, found differences in one or two bands and interpreted this finding as evidence of a noticeable frequency of in vivo transposition of this insertion element. RFLP profiles of our isolates received from sputum at various phases of the treatment period differed from each other in 5 to 10 bands (Fig. 2). Identical RFLP profiles of two sputum isolates collected on consecutive days were indicative of a short-term stability of IS1245. Like many other authors (17, 27, 32, 41, 43), we observed a considerable heterogeneity among our isolates and clones in tests with the IS1245 probe.
IS1311 RFLP types obtained by hybridization of isolates from HIV-positive patients in Spain were characterized by considerable heterogeneity. RFLP analyses of 75 isolates yielded more than 19 types (43). Our analyses of all 36 clones derived from the six sputum isolates from an HIV-negative patient and six clones from a dust isolate yielded only four RFLP types (Fig. 3). Our finding is consistent with the low discrimination potency of IS1311 reported earlier by Roiz et al. (43) and O'Grady et al. (33), who detected only six to eight copies of this transposon in the genome.
Since the isolates at the beginning and towards the end of the disease differed in genotypic and phenotypic characteristics, we can assume that the patient was infected by more than a single clone (as shown in IS1245 RFLP subtypes A1, A2, A3, and A4 present in two clusters, I and III, and subtypes E1, E2, E3, and E4, present in two clusters, I and II). It has been demonstrated in Brazil that, unlike M. tuberculosis, infections by the facultatively pathogenic M. avium complex agents can be polyclonal; this fact can cause therapeutic problems if isolates differing in drug resistance are involved (44).
The hypothesis of a concurrent or consecutive infection is supported by the finding of lung lesions in our patient which were indicative of inhalation of the agent from an environmental source. Along with deglutition, inhalation is considered the most frequent route of human infections by M. avium complex isolates (7, 10, 54). One of the isolates from the dust collected in the patient's home had the same genotype, dnaJ+ IS901− IS1311+ IS1245+, as the sputum isolates. Isolates of this genotype are also most frequently isolated from environmental samples and from infected swine and humans (6, 17, 24). Our finding allows the conclusion that home dust was the major risk factor for our patient (Table 2).
Japanese authors demonstrated M. avium complex isolates in 68.0% of dust samples tested (21). Our findings were much lower; the presence of M. avium complex isolates was demonstrated in only 2 (4.2%) of the 48 dust samples tested. Our conclusion is also supported by the identification of serotype 9 in one of the dust isolates and in four sputum isolates collected during a 4-month period of M. avium complex shedding in the sputum. On the other hand, dust could be contaminated by aerosols from the patient's cough.
Another possible and highly hazardous source in the case under study was soil with which the patient came into contact frequently during repotting of flowers and work in her garden. M. avium complex isolates detected in soil can originate from the excrement of domestic animals (4, 5, 9). Our serotype 2, genotype IS901+ IS1311+ IS1245+ isolate (Table 2) was probably of avian origin, because it is known most frequently to occur in birds (36). The serotype 6 sputum isolates may have originated from soil. Reznikov et al. (40) and Masaki et al. (28, 29) reported the isolation of this serotype of M. avium from soil samples collected in the vicinity of pig farms.
The most convincing proof supporting the hypothesis of concurrent or consecutive infection was a considerable heterogeneity found in RFLP analyses of clones derived from the sputum isolates (Fig. 2). By Gel Compar analysis, no obvious similarity was observed between RFLP types and corresponding RFLP subtypes (Fig. 2 and 4). The RFLP subtypes A1, A2, A3, and A4 were presented in different clusters (I and III) and clades (Ia and Ib), which confirmed their nonclonal origin. RFLP profile A can be extrapolated as a mixture of individual A1, A2, A3, and A4 RFLP profiles. The same situation was observed for RFLP subtypes derived from RFLP type E. The corresponding RFLP subtypes, B3, E1, E2, E3 and E4, were presented in clusters I and II and clades Ia and Ib. The relatively more genetically homogeneous groups were formed by RFLP subtypes derived from RFLP types B and F.
Our results are consistent with those of Pestel-Caron and Arbeit (38), who observed differences in one or two fragments among the IS1245 RFLP profiles of separate M. avium complex colonies grown in a primary culture from a patient affected by a polyclonal infection. Our results indicate that none of the isolates of exact serotype or RFLP profile persisted in the patient throughout the treatment period. It should be noted that the individual RFLP subtypes were not always replaced by new ones and that the B and E RFLP types included an A1 and B3 subtype, respectively. It could be supported by the demonstration of clones of mixed serotype 6/9 during the whole treatment period (Table 1). We conclude from the established wide array of serotypes and some different RFLP subtypes obtained from clones that the patient was infected by more than a single mycobacterial clone of M. avium.
The hypothesis of a reactivation of endogenous infection manifested by aggravation of clinical symptoms suppressed initially due to inappropriate drug treatment can be rejected by virtue of changing IS1245 RFLP types or subtypes. RFLP profiles of the individual isolates and clones differed from each other at the beginning of the disease and changed subsequently (Table 1). Also phenotypic characteristics were variable, specifically the MICs of selected antituberculosis drugs. Monitoring during the 5-month treatment period revealed increasing resistance of all the clones tested to several drugs (Table 1). No relation between RFLP profile and MIC was observed. Among-clone differences in MIC complicated the treatment of our patient (34). A similar problem was reported by Saad et al. (44).
The hypothesis of microevolution of isolates would be true if progressive changes of genotypic characteristics of the individual isolates/clones were demonstrated. Such progressive changes could be traced particularly within cluster I or its clades Ia and Ib, as was shown in IS1245 RFLP subtypes A1, A2, A3, and A4 present in two clusters, I and III, and two clades, Ia and Ib (Table 1, Fig. 4). The hypothesis cannot be unambiguously rejected on the basis of MICs, either.
Although a progressive selection of isolates showing susceptible pattern a could be traced, such putative microevolution could not significantly influence the resistance against drugs used within the study, because most of the clones were already resistant. No links among the individual RFLP spectra of isolates and clones that would unambiguously corroborate the hypothesis of transformation of one RFLP subtype or clone into another due to a simple transposition or site-specific recombination were demonstrable.
FIG. 5.
IS1245 RFLP subtypes of a dust isolate and its clones. Sizes are shown in kilodaltons.
Acknowledgments
We thank Tim Bull from St. George's Hospital in London (United Kingdom) for his stimulating, thought-provoking suggestions and comments and Marcela Fisakova and Zdenka Rozsypalova from the Veterinary Research Institute in Brno (Czech Republic) for their excellent technical assistance. Plasmid pMA12 with the cloned IS1245-specific fragment was kindly provided by Pieter Overduin of the RIVM, Bilthoven, The Netherlands.
This study was supported by the National Agency for Agricultural Research (grant no. QC 0195) and Research Project MZE-M03-99-01 of the Ministry of Agriculture of the Czech Republic and by the Programmes from Brussels, EC nos. FAIR6-CT98-4373 and QLK2-CT-2000-00928.
REFERENCES
- 1.Bartu, V. 1998. Pulmonary and extrapulmonary mycobacterioses. Epidemiol. Mikrobiol. Imunol. 47:20-22. (In Czech.) [PubMed] [Google Scholar]
- 2.Bauer, J., and A. B. Andersen. 1999. Stability of insertion sequence IS1245, a marker for differentiation of Mycobacterium avium strains. J. Clin. Microbiol. 37:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, J., A. B. Andersen, D. Askgaard, S. B. Giese, and B. Larsen. 1999. Typing of clinical Mycobacterium avium complex strains cultured during a 2-year period in Denmark by using IS1245. J. Clin. Microbiol. 37:600-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerwerth, W., and U. Kessel. 1976. Aviäre Mykobakterien im Kot von Wild- und Zoovögeln. Prax. Pneumol. 30:374-377. [PubMed] [Google Scholar]
- 5.Beerwerth, W., and J. Schürmann. 1969. Zür Ökologie der Mykobakterien. Erste Abt. Orig. Bd. 211:58-69. [PubMed] [Google Scholar]
- 6.Bono, M., T. Jemmi, C. Bernasconi, D. Burki, A. Telenti, and T. Bodmer. 1995. Genotypic characterization of Mycobacterium avium strains recovered from animals and their comparison to human strains. Appl. Environ. Microbiol. 61:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin, D. P., P. C. Hopewell, D. M. Yajko, E. Vittinghoff, C. R. Horsburgh, Jr., W. K. Hadley, E. N. Stone, P. S. Nassos, S. M. Ostroff, and M. A. Jacobson. 1994. Mycobacterium avium complex in the respiratory or gastrointestinal tract and the risk of M. avium complex bacteremia in patients with human immunodeficiency virus infection. J. Infect. Dis. 169:289-295. [DOI] [PubMed] [Google Scholar]
- 8.Collins, D. M., S. Cavaignac, and G. de Lisle. 1997. Use of four DNA insertion sequences to characterize strains of the Mycobacterium avium complex isolated from animals. Mol. Cell. Probes 11:373-380. [DOI] [PubMed] [Google Scholar]
- 9.Costallat, L. F., A. F. P. De Castro, A. C. Rodrigues, and F. M. Rodrigues. 1977. Examination of soils in the Campinas rural area for microorganisms of the Mycobacterium avium-intracellulare-scrofulaceum complex. Aust. Vet. J. 53:349-350. [DOI] [PubMed] [Google Scholar]
- 10.Damsker, B., and E. J. Bottone. 1985. Mycobacterium avium-Mycobacterium intracellulare from the intestinal tract of patients with the acquired immunodeficiency syndrome: concepts regarding acquisition and pathogenesis. J. Infect. Dis. 151:179-181. [DOI] [PubMed] [Google Scholar]
- 11.Dautzenberg, B. 1996. Rifabutin in the treatment of Mycobacterium avium complex infection: experience in Europe. Clin. Infect. Dis. 1:33-36. [DOI] [PubMed] [Google Scholar]
- 12.Dvorska, L., I. Parmova, M. Lavickova, J. Bartl, V. Vrbas, and I. Pavlik. 1999. Isolation of Rhodococcus equi and atypical mycobacteria from lymph nodes of pigs and cattle in herds with the occurrence of tuberculoid gross changes in the Czech Republic over the period 1996-1998. Vet. Med. Czech 44:321-330. [Google Scholar]
- 13.Falkinham, J. O. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, O., L. Matlova, J. Bartl, L. Dvorska, I. Melicharek, and I. Pavlik. 2000. Findings of mycobacteria in insectivores and small rodents. Folia Microbiol. 45:147-152. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, O., L. Matlova, L. Dvorska, P. Svastova, J. Bartl, I. Melicharek, R. T. Weston, and I. Pavlik. 2001. Diptera as vectors of mycobacterial infections in cattle and pigs. Med. Vet. Entomol. 15:208-211. [DOI] [PubMed] [Google Scholar]
- 16.Grange, J. M. 1996. Mycobacteria and human disease, 2nd ed. Arnold, London, England.
- 17.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hejlicek, K., M. Kubin, and F. Treml. 1997. The significance of ovarian tuberculosis in small poultry breedings for the health status of breeders. Stud. Pneumol. Phtiseol. 57:179-182. (In Czech.) [Google Scholar]
- 19.Horsburgh, C. R., Jr., D. P. Chin, D. M. Yajko, P. C. Hopewell, P. S. Nassos, E. P. Elkin, W. K. Hadley, E. N. Stone, E. M. Simon, P. Gonzalez, S. Ostroff, and A. L. Reingold. 1994. Environmental risk factors for acquisition of Mycobacterium avium complex in persons with Human Immunodeficiency Virus infection. J. Infect. Dis. 170:362-367. [DOI] [PubMed] [Google Scholar]
- 20.Horvathova, A., J. Kazda, J. Bartl, and I. Pavlik. 1997. Occurrence of conditionally pathogenic mycobacteria in the environment and their impact on living organisms Vet. Med. (Praha) 42:191-212. (In Slovak.) [PubMed] [Google Scholar]
- 21.Ichiyama, S., K. Shimokata, and M. Tsukamura. 1988. The isolation of Mycobacterium avium complex from soil, water, and dusts. Microbiol. Immunol. 32:733-739. [DOI] [PubMed] [Google Scholar]
- 22.Kaustova, J. 1997. Quantitative micromethod for drug susceptibility testing of mycobacteria in Sula's medium. Klin. Mikrobiol. Infect. Lek. 3:115-124. [Google Scholar]
- 23.Kazda, J. 2000. The ecology of the mycobacteria, 1st ed. Kluwer Academic Publishers, London, England.
- 24.Komijn, R. E., P. E. de Haas, M. M. Schneider, T. Eger, J. H. Nieuwenhuijs, R. J. van den Hoek, D. Bakker, F. G. van Zijderveld, and D. van Soolingen. 1999. Prevalence of Mycobacterium avium in slaughter swine in The Netherlands and comparison of IS1245: restriction fragment length polymorphism patterns of porcine and human isolates. J. Clin. Microbiol. 37:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubin, M. 1996. Tuberculosis semper viva. Cas. Lek. Ces. 135:39-42. (In Czech.) [PubMed] [Google Scholar]
- 26.Kubin, M., B. Burianova, L. Mezensky, M. Slosarek, and M. Turzova. 1986. Diagnosis of mycobacterial infections, p. 32- 42. In J. Schindler et al. (ed.), Microbiology methods, part 3, 1st ed. Avicenum, Prague, Czech Republic.
- 27.Kunze, Z. M., F. Portales, and J. J. McFadden. 1992. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J. Clin. Microbiol. 30:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masaki, S., K. Shimizu, N. Cho, and T. Hirose. 1981. Isolation of mycobacteria from lymph nodes of pig and their environment. J. Jpn. Vet. Med. Assoc. 44:213-221. [DOI] [PubMed] [Google Scholar]
- 29.Masaki, S., T. Konishi, G. Sugimori, A. Okamoto, Y. Hayashi, and F. Kunze. 1989. Plasmid profiles of Mycobacterium avium complex isolated from swine. Microbiol. Immunol. 33:429-433. [DOI] [PubMed] [Google Scholar]
- 30.Matlova, L., O. Fischer, J. Kazda, J. Kaustova, J. Bartl, A. Horvathova, and I. Pavlik. 1998. Occurrence of mycobacteria in invertebrates and poikilothermic animals and their role in man and other animals. Vet. Med. Czech 43:115-132. (In Czech.) [Google Scholar]
- 31.Musial, C. A., L. S. Tice, L. Stockman, and G. D. Roberts. 1988. Identification of mycobacteria from culture by with the Gen-Probe rapid diagnostic system for Mycobacterium avium complex and Mycobacterium tuberculosis complex. J. Clin. Microbiol. 26:2120-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai, R., S. Takewaki, A. Wada, K. Okuzumi, A. Tobita, and A. Ohkubo. 1990. Development of rapid detection method for mycobacteria with PCR. J. Med. Technol. 38:1247-1252. (In Japanese.) [PubMed] [Google Scholar]
- 33.O'Grady, D., O. Flynn, E. Costello, F. Quigley, A. Gogarty, J. McGuirk, J. O'Rourke, and N. Gibbons. 2000. Restriction fragment length polymorphism analysis of Mycobacterium avium isolates from animal and human sources. Int. J. Tuberc. Dis. 4:278-281. [PubMed] [Google Scholar]
- 34.Ostadal, O., J. Kaustova, M. Rehulka, and J. Bystron. 1999. Avian mycobacteriosis. Remedia Klin. Mikrobiol. 3:22-26. (In Czech.) [Google Scholar]
- 35.Pavlas, M., and V. Patlokova. 1985. Occurrence of mycobacteria in sawdust, straw, hay and their epizootological significance. Acta Vet. Brno 54:85-90. [Google Scholar]
- 36.Pavlik, I., P. Svastova, J. Bartl, L. Dvorska, and I. Rychlik. 2000. Relationship between IS901 in the Mycobacterium avium complex strains isolated from birds, animals, humans and environment and virulence for poultry. Clin. Diagn. Lab. Immunol. 7:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlik, I., J. Bartl, L. Dvorska, P. Svastova, M. Havelkova, and M. Slosarek. 1999. Current health importance and identification of the Mycobacterium avium complex in man and animals. Remedia Klin. Mikrobiol. 3:5-12. (In Czech.) [Google Scholar]
- 38.Pestel-Caron, M., and R. D. Arbeit. 1998. Characterization of IS1245 for strain typing of Mycobacterium avium. J. Clin. Microbiol. 36:1859-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piening, C., W. Anz, and G. Meissner. 1972. Serotyp Bestimmungen und ihre Bedeutung für epidemiologische Untersuchungen bei der Schweinetuberkulose in Schleswig-Holstein. Dte. Tierärztl. Wochenschr. 79:85-93. [PubMed] [Google Scholar]
- 40.Reznikov, M., and D. J. Dawson. 1971. Serological investigation of strains of Mycobacterium intracellulare (Battey bacillus) isolated from house-dusts. Med. J. Aust. 58:682-683. [DOI] [PubMed] [Google Scholar]
- 41.Ritacco, V., K. Kremer, T. van der Laan, J. E. Pijnenburg, P. E. W. de Haas, and D. van Soolingen. 1998. Use of IS901 and IS1245 in RFLP typing of Mycobacterium avium complex: relatedness among serovar reference strains, human and animal isolates. Int. J. Tuberc. Lung. Dis. 2:242-251. [PubMed] [Google Scholar]
- 42.Roberts, M. C., C. H. McMillan, and M. B. Coyle. 1987. Whole chromosomal DNA probes for rapid identification of Mycobacterium tuberculosis and Mycobacterium avium complex. J. Clin. Microbiol. 25:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roiz, M. P., E. Palenque, C. Gerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saad, M. H. F., M. A. Telles, F. Porfirio, L. Ferrazoli, L. S. Fonseca, W. Johnson, Jr., and L. W. Riley. 2000. Multiple isolates from AIDS patients: Aspects of an analysis by a genotypic marker and antimicrobial susceptibilities variations. Mem. Inst. Oswaldo Cruz 95:729-732. [DOI] [PubMed] [Google Scholar]
- 45.Saito, H., H. Tomioka, K. Sato, H. Tasaka, M. Tsukamura, F. Kuze, and K. Asano. 1989. Identification and partial characterization of Mycobacterium avium and Mycobacterium intracellulare by using DNA probes. J. Clin. Microbiol. 27:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito, H., H. Tomioka, K. Sato, H. Tasaka, and D. Dawson. 1990. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 28:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Süssland, Z., and V. Hrdinova. 1976. Use of rapid agglutination in the serotyping of the Mycobacterium avium-intracellulare complex. Vet. Med. (Praha) 21:209-213. (In Czech.) [PubMed] [Google Scholar]
- 48.Thoen, C. O., and J. H. Steele. 1995. Mycobacterium bovis infection in animals and humans. Iowa State University Press, Ames, Iowa.
- 49.Thorel, M. F., H. F. Huchzermeyer, and A. L. Michel. 2001. Mycobacterium avium and Mycobacterium intracellulare infection in mammals. Rev. Sci. Tech. 20:204-218. [DOI] [PubMed] [Google Scholar]
- 50.van Soolingen, D., P. E. W. de Haas, P. W. M. Hermans, and J. D. A. van Embden. 1993. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 51.Wayne, L. G., and G. P. Kubica. 1986. Family Mycobacteriaceae Chester 1897, 63AL. In P. H. Sneath et al. (ed.), Bergey's manual of systematic bacteriology. Williams and Wilkins, Baltimore, Md.
- 52.Whipple, D. L., and R. S. Merkal. 1985. Procedures for the field and laboratory processing of fecal specimens for the isolation of Mycobacterium paratuberculosis. Proc. U.S. Anim. Health Assoc. 89:475-479. [Google Scholar]
- 53.Wolinsky, E., and W. B. Schaefer. 1973. Proposed numbering scheme for mycobacterial serotypes by agglutination. Int. J. Syst. Bacteriol. 23:182-183. [Google Scholar]
- 54.Yajko, D. M., D. P. Chin, P. C. Gonzales, P. S. Nassos, P. C. Hopewell, A. L. Reingold, C. R. Horsburgh, M. A. Yakrus, S. M. Ostroff, and W. K. Hadley. 1995. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 9:176-182. [PubMed] [Google Scholar]