Abstract
Genotypic diversity among 26 isolates of Bartonella bacilliformis obtained from different areas of Peru, and at different times, was assessed by comparison of DNA sequences derived from 16S-23S ribosomal DNA intergenic spacer regions (ISR) and a citrate synthase gene (gltA) fragment and by amplified fragment length polymorphism (AFLP) analysis. gltA comparison divided the isolates into two groups, whereas ISR comparison revealed six sequences. AFLP analysis using a selective primer delineated five profiles that correlated well with those obtained by sequence comparison. Combination of all three data sets divided the isolates into six genotypes. One of these genotypes was common to isolates collected from a large area in western Peru that corresponded to the region of endemicity for bartonellosis; however, isolates belonging to two other genotypes were also found within this region. Two of these genotypes were found in isolates isolated more than 35 years apart. The remaining three genotypes were each specifically associated with three outbreaks of bartonellosis that have recently occurred in areas where the disease had not previously been recognized. Demonstration of the unique nature of these isolates indicates that the outbreaks with which they were associated did not result from the introduction of disease by individuals who acquired their infection in the recognized region of endemicity. The sources of these outbreaks remain unknown. A consensus approach to bacterial typing using comparative sequence analysis of multiple genetic loci and the pan-genomic sampling of AFLP appears to offer a well-supported assessment of B. bacilliformis diversity, and the genotypic differences identified appear to have epidemiological significance.
The recent emergence and reemergence of Bartonella henselae and Bartonella quintana as organisms of medical importance in Europe and the United States have overshadowed the long-established role of Bartonella bacilliformis as the agent of bartonellosis in Peru, Colombia, and Ecuador. However, in terms of morbidity and mortality, B. bacilliformis remains the biggest threat to human health of all the Bartonella species. Overt infection manifests in one of two quite distinct ways, either as a severe acute-onset anemia (Oroya fever) or as angiogenic skin lesions termed verrugas (verruga peruana). Although the latter manifestation can be visually quite alarming, the condition is benign and self-limiting. Conversely, Oroya fever is often life threatening as, if untreated, virtually all erythrocytes can become infected by bacteria (13, 24). Immunocompromise induced by this anemia renders the afflicted highly susceptible to opportunistic secondary infections, and it is these that often kill (13, 17, 24).
Although most text books state that, classically, verruga peruana occurs in patients who have suffered and recovered from Oroya fever, this progression is, in fact, just one of several possible disease courses. Severe Oroya fever is rarely seen among the population in regions of endemicity, but verruga peruana is common (17). In areas of nonendemicity, Oroya fever appears to be more common (13, 24), although this is not always the case (15). The basis for these observations has been proposed as either variation in host immunity (24) (although the immunological parameters of patients have never been assessed) or variation in pathogenicity between B. bacilliformis strains (15). In addition to these overt clinical manifestations, asymptomatic infections have also been frequently encountered, supporting the idea that humans are maintenance hosts for the bacterium (13, 15, 25). A recent serological survey of healthy volunteers living in an area of bartonellosis endemicity found that 45% of 387 people tested were seropositive for Bartonella antibodies (5), further corroborating the view that infection by B. bacilliformis is common.
Outbreaks of bartonellosis not only continue in recognized regions of endemicity but are also now being reported with increasing frequency in areas where the disease has not previously been encountered (8, 15). The public health impact of bartonellosis in these areas is at least as significant as it is in regions of endemicity. For example, a survey of 554 individuals (55% of the population) living in a new focus of bartonellosis in the north of Peru found that 77% had serological evidence of B. bacilliformis infection (15). Furthermore, the proportion of this community who suffered overt symptoms was remarkably high, with 14% developing Oroya fever and 18% developing verruga peruana.
Despite being recognized for almost 100 years, B. bacilliformis remains little studied, and fundamental issues regarding the epidemiology of bartonellosis remain unanswered. It is therefore extremely timely to exploit some of the new microbiological methods described for the characterization and epidemiological surveillance of B. henselae and B. quintana for the study of B. bacilliformis. For the first time, this report compares the genotypes of clinical isolates of B. bacilliformis obtained from patients in different regions of Peru and on different dates. Bacterial typing was achieved using a combination of established sequence comparison-based methods (3, 19, 20) and a sensitive PCR-based pan-genomic sampling method, amplified fragment length polymorphism (AFLP) analysis (23).
MATERIALS AND METHODS
Terminology.
The following definitions of the terms isolate and strain proposed by Struelens and colleagues (22) are used throughout the text: “isolate,” a population of microbial cells in pure culture derived from a single colony on an isolation plate and characterized by identification to the species level; “strain,” an isolate or group of isolates exhibiting phenotypic and/or genotypic traits which are distinctive from those of other isolates of the same species.
Bacterial isolates.
A total of 26 isolates of B. bacilliformis were included in the study. The sources of these isolates, together with their date of isolation, are presented in Table 1. The isolates CON600-1 and Cond044 were kindly provided by Larry Laughlin and Judith Chamberlin of the Uniformed Services University of the Health Sciences, Bethesda, Md.
TABLE 1.
B. bacilliformis isolates used in this study and results of genotypic assessments
| Isolate designation | Locationa where bartonellosis occurred | Date of isolationb | Patient characteristic
|
ISR type | AFLP type | ||
|---|---|---|---|---|---|---|---|
| Age (yr) | Genderc | Clinical statusd | |||||
| KC583T | Huarochiri (p), Lima (2,738 m) | Early 1960s | NKe | NK | NK | A | 1 |
| KC584 | Churcampa (p), Huancavelica (3,262 m) | Early 1960s | NK | NK | NK | A | 1 |
| LA6.2 | Huayllacayan (d), Bolognesi (p), Ancash (3,256 m) | 19/12/1990 | NK | NK | OF | E | 5 |
| LA6.3 | Huayllacayan, Bolognesi, Ancash | 23/12/1990 | NK | NK | OF | E | 5 |
| Monz-269 | Monzon (d), Huamalies (p), Huanuco (1,800 m) | 17/02/1998 | 43 | F | OF | C | 4 |
| Acoch- 812 | Acochaca (d), Asuncion (p), Ancash (3,350 m) | 16/03/1998 | NK | F | OF | A | 1 |
| CON600-1 | Huaylas (d), Huaylas (p), Ancash (2,721 m) | 13/06/1997 | 8 | F | OF | B | 2 |
| Cond044 | Huaylas, Huaylas, Ancash | 13/06/1997 | 10 | F | AS | A | 1 |
| Cusco5 | Urubamba (d), Urubamba (p), Cusco (2,871 m) | 4/1998 | 5 | M | OF | D | 3 |
| Cusco8 | Urubamba, Urubamba, Cusco | 5/1998 | 28 | M | OF | D | 3 |
| Cusco14 | Urubamba, Urubamba, Cusco | 4/1998 | 2 | M | OF | D | 3 |
| Cusco407 | Urubamba, Urubamba, Cusco | 4/1998 | NK | M | AS | D | 3 |
| Cusco-Ana | Urubamba, Urubamba, Cusco | 4/1998 | 10 | M | OF | D | 3 |
| Sih-Ism | Sihuas (d), Sihuas (p), Ancash (2,716 m) | 5/1999 | 28 | M | OF | A | 1 |
| T2 | Huarez (p), Ancash (3,136 m) | 2/1999 | NK | NK | OF | A | 1 |
| Luc-Uba | Piscobamba (d), Mariscal Luzuriaga (p), Ancash (3,281 m) | 5/1999 | 4 | F | OF | E | 5 |
| Hua-Chu | Sangallaya (d), Huarochiri (p), Lima (2,738 m) | 5/1999 | 16 | M | OF | A | 1 |
| Hua-Mar | Sangallaya, Huarochiri, Lima | 6/1999 | 2 | M | OF | A | 1 |
| Hua-Rub | Sangallaya, Huarochiri, Lima | 4/1999 | 23 | M | OF | A | 1 |
| Hua-Nol | Sangallaya, Huarochiri, Lima | 7/1999 | 50 | M | OF | A | 1 |
| ER-Yal | Pisuquia (d), Luya (p), Amazonas (2,000 m) | 2/1999 | NK | M | OF | F | 3 |
| ER-Tej | Pisuquia, Luya, Amazonas | 7/1999 | 38 | M | OF | F | 3 |
| ER-Cha | Pisuquia, Luya, Amazonas | 7/1999 | 16 | M | OF | F | 3 |
| NCTC12134 | NK | 1949 | NK | NK | NK | B | 2 |
| NCTC12135 | NK | 1941 | NK | NK | NK | B | 2 |
| NCTC12136 | NK | 1957 | NK | NK | NK | B | 2 |
Locations (all in Peru) are given, where possible, by district (d), province (p), and department (with altitude in parentheses).
Unless a general time name or year in indicated, dates are given as day/month/year or simply month/year.
M, male; F, female.
OF, Oroya fever; AS, asymptomatic.
NK, not known.
All isolates were cultivated on Columbia blood agar incorporating 10% whole horse blood incubated at 30°C for, typically, 5 days in a moist atmosphere. Prior to the sequence-based methods described below, the newly described isolates were identified as B. bacilliformis on the basis of their colonial appearance, their microscopic morphology, their ability to grow at 30°C but not at 37°C, and their serological reactivity with polyvalent B. bacilliformis antisera (4).
DNA extraction, PCR amplifications, and nucleotide sequence determinations.
Crude DNA extracts, suitable for use as templates in PCR-based amplification reactions, were prepared from sweeps of bacterial colonies using the QIAamp DNA extraction kit (Qiagen, Crawley, West Sussex, United Kingdom) by following the manufacturer's instructions. Each extract was then incorporated into two previously described PCRs. A citrate synthase gene (gltA) fragment was amplified using a Bartonella-specific PCR incorporating the primer pair 443f-1137r (3, 19) as previously described (3). The complete intergenic spacer region (ISR) between the 16S and 23S rRNA genes was amplified using a broad-spectrum PCR incorporating the primer pair 16S1386f-23S115r (7, 16). Each 50-μl reaction mixture contained 2 μl of each primer at 10 pmol/μl, 2 μl of DNA extract, 25 μl of 2× extensor PCR mastermix (ABgene, Epsom, United Kingdom), and 19 μl of sterile distilled water. The thermal cycle consisted of an initial denaturation at 96°C for 3 min, followed by 40 cycles of denaturation at 96°C for 10 s, primer annealing at 48°C for 20 s, and primer extension at 72°C for 40 s. The program was completed with a stage at 72°C for 6 min. The success of each PCR was gauged by UV visualization of ethidium bromide-stained 1% agarose gels on which 10 μl of each amplification product had been electrophoretically resolved.
Amplification products were purified for use as templates in nucleotide base sequencing reactions using the QIAquick purification kit (Qiagen) according to the manufacturer's instructions. Cycle sequencing reactions were prepared using the dRhodamine terminator cycle sequencing ready reaction kit (Perkin-Elmer, Warrington, United Kingdom) according to the manufacturer's instructions. Reactions for gltA analysis incorporated the primers used in the initial amplification reactions. Reactions for ISR analysis incorporated the primers used in initial PCRs together with the previously described QHVE1 and QHVE3 (20). The thermal program employed for all sequencing reactions was 30 cycles of 95°C for 20 s, 50°C for 10 s, and 60°C for 4 s. Reaction products (10 μl) were mixed with 74 μl of 70% ethanol-0.5 M MgCl2, and, after being held for 1 h at 4°C, precipitating DNA was collected by centrifugation at 14,000 × g for 12 min. DNA pellets were dried and resuspended in 3 μl of a 5:1 mixture of formamide-1% dextran blue and 0.025 M EDTA. Purified products were resolved on a 5% denaturing acrylamide gel (Long Ranger Singel 377-36; FMC Bioproducts, Rockland, Maine), and sequence data were captured and prepared using an ABI 377 automated sequencer and related software (Perkin-Elmer).
AFLP analysis.
Genomic DNA suitable for use as a template in the PCR was extracted with the Nucleon genomic DNA extraction kit (GeneSys Biotech Ltd., Coatbridge, Strathclyde, United Kingdom). DNA concentration was determined spectrophotometrically by measuring the absorbance at 260 nm. The AFLP method used here is based on the Standard European Working Group on Legionella Infections protocol (9) but uses the selective primer PstI-C. Restriction-ligation reactions were performed at 37°C for 3 h in a total volume of 20 μl. Each mix comprised approximately 1.5 μg of genomic DNA, 200 ng of each adapter-oligonucleotide (AFLP-LG1, 5′-CTCGTAGACTGCGTACATGCA-3′; AFLP-LG2, 5′-TGTACGCAGTCTAC-3′), 20 U of PstI (Boehringer Mannheim GmbH, Mannheim, Germany), 1 U of T4 DNA ligase (Boehringer), and 1× ligation buffer (10× ligation buffer is 660 mM Tris [pH 7.5], 50 mM MgCl2, 10 mM dithiothreitol, 10 mM ATP; Boehringer). Prior to the PCR, tagged DNA fragments were precipitated using a final concentration of 2.5 M ammonium acetate in 100 μl and an equal volume of absolute ethanol. After incubation for 5 min at room temperature, centrifugation was carried out at 12,000 × g for 10 min, and the pellet washed once with 70% ethanol. The precipitate was air dried and resuspended in 100 μl of TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). Before the PCR a 1-in-100 dilution of the resuspended DNA was prepared, and 5 μl of this dilution was used as a template DNA in the PCR. PCR was performed in a reaction mixture of 25 μl using the Ready-To-Go PCR beads (0.5-ml format; Amersham Pharmacia Biotech). Each reaction mixture comprised template DNA and 75 ng of selective primer (AFLP-PstI-C, 5′-GACTGCGTACATGCAGC-3′), and the magnesium concentration was adjusted to 2.5 mM MgCl2. Amplification was performed using the following parameters: initial denaturation at 94°C for 4 min followed by 33 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and elongation at 72°C for 2.5 min. Amplified products (20 to 40% of the reaction mix; i.e., 5 to 10 μl) were separated by electrophoresis on 1.5% agarose (UltraPure agarose; Life Technologies) gels in 1× TBE (0.089 M Tris-borate, 1 mM EDTA) for 4 h at 3.5 V/cm. The MBI Fermentas (Sunderland, United Kingdom) Ladder mix (catalog no. SM0331) was used as molecular size markers (at 0.75 to 1.0 μg/lane). To aid normalization of the gels each sample lane was adjacent to a marker lane. Gels were stained with ethidium bromide (0.5 to 1.0 μg/ml) for 30 min and photographed under UV transillumination using Polaroid film 667 (Sigma).
Data analysis.
Black-and-white photographs of the gels were scanned with a ScanMaker (Microtek Lab) into Adobe Photoshop with 256 levels of grey and 400 dpi. The resultant TIFF files were then analyzed using BioNumerics software (version 2.5; Applied Maths BVBA, Sint-Martens-Latem, Belgium). AFLP patterns were analyzed using the clustering (curve based) option (Pearson correlation) with the unweighted pair group method with averages method. Percentage similarity thresholds were used to divide the output from the dendrograms into types.
ISR sequences were assembled using Align Plus 4 (Scientific and Educational Software, Durham, N.C.). An alignment of complete sequence data was made, and a matrix of uncorrected distances was calculated (BioNumerics). A relatedness dendrogram was derived using neighbor-joining analysis, and bootstrapping of this data was performed using 1,000 replications (BioNumerics).
Statistical methods.
Determination of the limitation of the sample size in estimating the diversity of B. bacilliformis genotypes present in the areas surveyed was based on a binomial probability distribution (26). From this analysis, the probability that the new foci of bartonellosis were caused by strains extant at an undetected, low prevalence in the zone of endemicity could also be estimated.
Nucleotide sequence accession numbers.
The five new ISR sequences have been deposited in GenBank under the indicated accession numbers: LA6.3, AJ422181; Monz-269, AJ422182; CON600-01, AJ422179; Cusco407, AJ422180; ER-Yal, AJ422178.
RESULTS
gltA analysis.
Partial gltA sequences of about 700 bp were obtained for all isolates. Comparison of these sequences revealed that the isolates possessed one of two different sequences. The two isolates obtained from the Huayllacayan valley in 1990 and the Luc-Uba isolate possessed an identical sequence that was 97% similar to that of the remaining 23 isolates, including the type strain, KC583. These two sequences had been deposited in GenBank prior to this study (3).
ISR analysis.
Complete ISR sequences were obtained for all 26 isolates of B. bacilliformis. All ISR sequences possessed a general structure very similar to that previously described for the ISR of the type strain. All contained genes encoding isoleucine tRNA and alanine tRNA that were separated by a centrally located variable region of approximately 120 bp and that were flanked by regions of approximately 300 bp. All sequences also contained previously described direct and inverted repeats (18). The sequences of both tRNA genes were identical in all isolates tested. Alignment and comparison of the 26 sequences revealed six variants. These different sequences varied in length as well as base sequence, ranging from 887 to 933 bp. The distribution of the six ISR genotypes among the 26 isolates is presented in Table 1, and the details of the differences between them are presented in Table 2. The shortest sequence was that obtained for the three isolates that possessed gltA heterogeneity. This sequence did not contain a 19-bp fragment present in the inter-tRNA gene region of the other five ISR genotypes. The longest sequences were those obtained from isolates from Urubamba and Monzon. These strains contained a 25-bp fragment lying between the two inverted repeat sequences towards the 3′ end of the ISR which was not present in other isolates tested. Compared to the ISR of the KC583 type strain, the isolates that bore the 25-bp insert and the isolates from Pisuquia also possessed two point insertions at 110 and 114 bp from the 5′ end of the ISR. On the same comparison, the ISR of the isolate CON600-01 and the three isolates obtained from the collection of the Institut Pasteur had a single base deletion at 267 bp from the 5′ end of the sequence. In addition to these insertion-deletion events, the six ISR genotypes differed by numerous base substitutions, as detailed in Table 2. Analysis of the single-nucleotide polymorphisms (SNPs) among the six ISRs indicated that isolates from Monzon, Urubamba, and Pisuquia differed from one another by fewer than 6 SNPs but differed from all other genotypes by more than 12 SNPs. Two of the genotypes associated with isolates from the zone of endemicity for bartonellosis differed by 6 SNPs but differed by more than 12 SNPs from other genotypes. The ISR of the gltA variant isolates differed by 20 or more SNPs compared with all other ISRs.
TABLE 2.
ISR sequence variation among the six B. bacilliformis genotypes identified
| Strain | ISR type | ISR length (bp) | Base(s) at indicated position(s) within intergenic spacer region (numbering relative to KC583 sequence)
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 85 | 101 | 110 | 114 | 131 | 189 | 200 | 207-208 | 216 | 225 | 263 | 269 | 436 | 443 | 447-465 | 471 | 505-506 | 508 | 522 | 620 | 625 | 743 | 743-744 | 748 | 753-755 | 794 | 812 | 881-883 | |||
| KC583 | A | 906 | C | G | C | G | G | TA | T | C | T | T | A | T | CCAGGCTTATCTTATAGTT | C | TG | C | A | G | G | T | A | GTG | G | C | CAG | |||
| CON600-1 | B | 905 | T | G | C | G | G | TA | T | C | C | A | A | CCAGGCTTATCTTATAGTT | T | TG | C | A | G | G | T | A | GTG | G | C | CAG | ||||
| Monz269 | C | 933 | T | G | A | T | C | T | A | TA | T | A | T | T | A | A | TGAGGCTTATCTTATAGTT | C | GA | G | G | G | G | T | TTTTAGAGAGTTTCTTTTAGGAGTT | A | GTT | G | C | CAT |
| Cusco-407 | D | 933 | T | G | A | T | C | T | A | TA | T | A | T | T | A | A | TGAGGCTTATCTTATAGTT | C | TG | C | G | G | G | T | TTTTAGAGAGTT-CTTTTAGGAGTT | A | GTT | G | C | CAG |
| LA6.3 | E | 887 | T | A | T | T | G | AC | A | A | T | C | G | A | C | TG | C | A | A | A | G | G | AGT | T | T | TGT | ||||
| ER-Yal | F | 908 | T | G | A | T | C | T | A | TA | T | A | T | T | A | A | TGAGGCTTATCTTATAGTT | C | GA | G | G | G | G | T | A | GTG | G | C | CAT | |
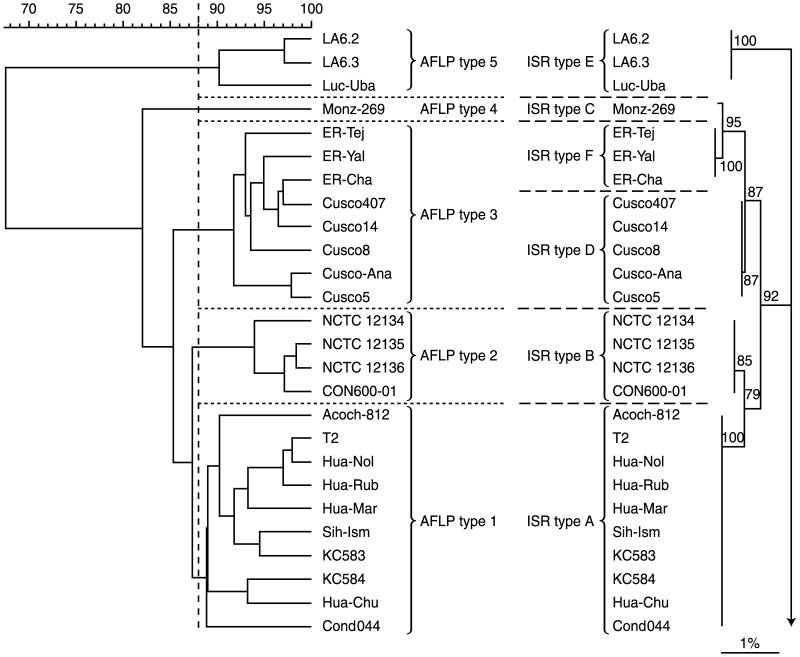
Analysis of the aligned sequences yielded a dendrogram (Fig. 1) in which (i) the gltA variant isolates formed an outgroup, (ii) isolates possessing the two other ISR sequences encountered in the bartonellosis zone of endemicity grouped together, and (iii) isolates possessing the three ISR sequences associated with recent outbreaks of bartonellosis in areas of nonendemicity grouped together. All of these groupings were well supported by bootstrap analysis.
FIG. 1.
Dendrograms based on AFLP analysis and ISR sequence similarities, showing relationships between B. bacilliformis isolates studied. The AFLP dendrogram (left) was constructed using Pearson analysis of normalized gels. The neighbor-joining dendrogram (right) shows the sequence similarities of ISRs. Bootstrap values, expressed as percentages of 1,000 replications, are given at branching points. The scale bar represents 1% sequence divergence. Strain designation, AFLP type, and ISR type are indicated.
AFLP analysis.
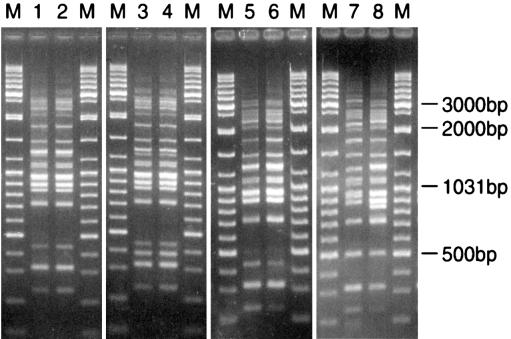
All isolates of B. bacilliformis included in this study yielded patterns using the AFLP protocol described above. Figure 2 shows examples of the patterns obtained following this protocol. Using a percentage similarity threshold (at or above which patterns were considered to belong to the same type) of 88%, five distinct AFLP types were found in the 26 isolates examined (Table 1; Fig. 1). Group 5 was outlying and included the three gltA sequence variant isolates. Groups 1 and 2, which together formed a cluster, included all other isolates from the bartonellosis region of endemicity. Group 3 included isolates from Urubamba and Pisuquia. Group 4, containing the isolate from Monzon, lay outside the two clusters defined above but clustered with them rather than with the outlying Group 5 isolates.
FIG. 2.
Examples of patterns obtained on agarose gel resolution of products derived from AFLP analysis of B. bacilliformis isolates. Lanes: 1, Hua-Nol; 2, Hua-Rub; 3, NCTC12134; 4, CON600-01; 5, Cusco-Ana; 6, Acoch-812; 7, Luc-Uba; 8, Monz-269; M, molecular size marker (GeneRuler; MBI Fermentas).
Epidemiological implications of genotyping results and statistical consideration of the limitations of sampling.
There was general congruence between ISR sequence- and AFLP-based genotypic assessments, although analysis of ISR delineated isolates from Urubamba and Pisuquia that belonged to the same AFLP type. Estimations of relative relatedness between genotypes made by each approach were slightly different in that ISR analysis indicated isolates from Monzon, Urubamba, and Pisuquia were all specifically and closely related, whereas AFLP analysis indicated that the isolate from Monzon was not specifically related to any other isolates studied. The most frequently encountered genotype was common to 10 isolates collected, over a 50-year period, from a large geographical area in central, western Peru, which corresponds to the principal region of endemicity for bartonellosis. However, isolates belonging to two other genotypes were also found within this region. One of these appeared to be very closely related to the most frequently encountered genotype, but the other, which comprised all isolates exhibiting gltA heterogeneity, was the most-divergent genotype, forming an outgroup in both ISR and AFLP assessments. In one district in the region of endemicity (Huaylas), two different genotypes were concurrently encountered. In one province of the region of endemicity (Huarochiri), the same genotype was encountered in the early 1960s and in 1999. Among the subjects from whom isolates were obtained, two were asymptomatic. The isolates obtained from both subjects were indistinguishable from isolates obtained from symptomatic patients in the same location. The remaining three genotypes were specifically related to three foci of bartonellosis that have recently occurred in regions of Peru not historically associated with the disease. Despite the incongruence of ISR- and AFLP-based cluster analysis discussed above, in no analysis was any of these three genotypes specifically related to those encountered in the zone of endemicity for bartonellosis.
A total of 17 B. bacilliformis isolates used in this survey were recovered from individuals living in the zone of endemicity for bartonellosis. Based on a binomial probability distribution analysis, only genotypes occurring at a prevalence of 16.2% or more among the B. bacilliformis population in the zone of endemicity would be 95% likely to be present in a sample of this size. However, the maximum probability that three genotypes with a cumulative prevalence of less than 16.2%, rather than more prevalent genotypes, were independently introduced from the zone of endemicity into the three sites where new foci of bartonellosis occurred was only 1.6 × 10−4 (maximum probability occurs when the three genotypes are at an equal [5.4%] prevalence).
DISCUSSION
All methods employed in this study allowed differentiation of B. bacilliformis isolates, although the sensitivity of gltA analysis was markedly less than that of the ISR and AFLP analyses. Comparison of partial gltA sequences demonstrated that all the isolates possessed either one of only two genotypes, both of which have been previously encountered. That gltA sequences remained very highly conserved among B. bacilliformis isolates was not surprising, as the stability of this gene among isolates of other Bartonella species has been previously reported (3). The difference between the two gltA genotypes is, however, surprisingly high (>3% dissimilarity). This degree of variation is close to that seen between the three subspecies of Bartonella vinsonii (14) and may therefore suggest that the B. bacilliformis isolates LA6.2, LA6.3, and Luc-Uba belong to a distinct taxon within the species. Genetic heterogeneity within B. bacilliformis has been demonstrated only once previously, when the sequence of a partial 16S rRNA gene PCR product obtained from a skin biopsy specimen of an Ecuadorian patient with verruga peruana was found to be only 96.5% similar to that of “known strains” (1). This degree of dissimilarity is, however, high for isolates of the same Bartonella species and suggests that the organism causing bartonellosis in the province of Manabi in Ecuador is a species different from B. bacilliformis.
As has been observed for other Bartonella species, ISR sequence comparison proved to be a markedly more sensitive approach to strain differentiation than gltA analysis (2, 20). The nature of the variation between the six different ISR sequences encountered was interesting. The presence of direct repeats in the ISR of other Bartonella species has been noted, and different isolates of the same species have been found to possess various numbers of these features (2). The application of an approach similar to the AFLP analysis described herein for the differentiation of Bartonella species has been previously reported (12). These studies highlighted the practicality of this approach, in relation to arbitrary PCR methods or macrorestriction analysis using pulsed-field gel electrophoresis (21), for the study of fastidious organisms such as bartonellae, and our use of the approach on B. bacilliformis supports these findings. The general consensus of pan-genomic sampling, as determined by AFLP analysis, and DNA sequence comparison is very satisfying considering the current concern regarding the relative value of these two approaches in phylogenetic, population genetic, or epidemiological studies (10). For such studies, the use of multilocus sequence comparison circumvents the potential pitfalls associated with relying solely on analysis of a single genetic locus in determining an accurate genetic identity for bacterial strains. However, reliance solely on comparison of sequences, even at several loci, ignores the importance of genomic rearrangement in shaping the genetic character of a bacterium. A combination of gene sequence analysis and an assessment of the overall genomic architecture of a bacterium should, therefore, serve as a practical means of sampling genotype, providing accurate and sensitive differentiation between strains.
The relatively small scale of this study dictates that any conclusions drawn from the comparison of B. bacilliformis genotypes and the epidemiological data available for the isolates should be considered with care. For example, the number of isolates surveyed from the zone of endemicity was such that we could only be confident of encountering genotypes that occurred at a prevalence of >16%. Nonetheless, our study yielded several observations from which noteworthy conclusions can be drawn. The three recent outbreaks of bartonellosis that occurred in Urubamba, Pisuquia, and Monzon, where the disease had not previously been known, were each caused by a unique genotype of B. bacilliformis and not by the genotype most often encountered in the region of Peru where bartonellosis is considered to be endemic. For each outbreak, all isolates obtained possessed the same genotype, indicating the epidemicity of the disease in each area. Even in the bartonellosis-endemic region, genotypic differences were observed. However, the most frequently encountered genotype was identified among isolates from patients from seven different locations in this region ranging from Churcampa in the south to Sihuas, which is over 600 km further north. Furthermore, as isolates collected more than 40 years ago also possessed this genotype, there appears to be a good degree of genetic stability among this bacterial population.
The basis for the apparent recent spread of bartonellosis into new areas of Peru remains unknown. Indeed, it may be foolhardy to assume that all new foci have appeared for the same reason. Our demonstration of genetic diversity among B. bacilliformis isolates associated with the new foci suggests that, rather than strains being introduced into the areas where these occurred, discrete populations of the species already existed. Such a hypothesis is supported by the demonstration that, even if the genotypes associated with new foci of infection were present at a low prevalence in the zone of endemicity, the likelihood that the new foci resulted from the chance introduction of these genotypes, and not those encountered at a higher prevalence, from the zone of endemicity was negligible. The possibility that B. bacilliformis populations can exist in areas where clinical bartonellosis is absent is intriguing and revives suspicion of nonhuman maintenance hosts. Equal intrigue surrounds the factors that underlie the appearance of bartonellosis in these areas. As recent vagaries in the epidemiologies of several infectious diseases in Peru and the South Pacific region have been associated with the El Niño phenomenon (6, 11), this climatic event may also have modulated changes in the epidemiology of bartonellosis.
The apparent variations in bartonellosis-related morbidity and mortality associated with different outbreaks of the disease have been previously highlighted. Specifically, Kosek and colleagues (15) hypothesized that the B. bacilliformis strain implicated in the disease focus they investigated may have possessed diminished virulence. Interestingly, the clinical observations supporting this hypothesis were also noted among patients in the Pisuquia outbreak, namely, that blood smears revealed an uncharacteristically low level of bacteremia and no patients developed a fulminant hemolytic disease (E. Sanchez, unpublished observations). Kosek and colleagues (15) suggested that strain differences may, in part, account for this disparity and called for further studies to address strain heterogeneity. Our study is the first to answer this request and to demonstrate that isolates derived from asymptomatic patients (CON600-01 and Cusco407) were indistinguishable from those causing overt disease.
Although this study has provided fundamental information regarding the epidemiology of bartonellosis in Peru, its findings have provoked questions rather than provided answers. Considerable work in this field is clearly necessary to improve our understanding of a disease of significant and increasing public health importance in the Andean region of South America.
Acknowledgments
We appreciate the technical assistance of J. Duncan of the Respiratory and Systemic Infection Laboratory. L. Laughlin and J. Chamberlin of the Uniformed Services University for the Health Sciences kindly provided clinical isolates and relevant epidemiological information. We are also grateful to S. Telfer for her help with statistical evaluation of the data and to M. Bennett and T. Harrison for review of the manuscript.
R. J. Birtles is supported by a Wellcome Trust Medical Microbiology Research Fellowship.
REFERENCES
- 1.Amano, Y., J. Rumbea, J. Knobloch, J. Olsen, and M. Kron. 1997. Bartonellosis in Ecuador: serosurvey and current status of cutaneous verrucous disease. Am. J. Trop. Med. Hyg. 57:174-179. [DOI] [PubMed] [Google Scholar]
- 2.Birtles, R. J., S. Hazel, K. Bown, D. Raoult, M. Begon, and M. Bennett. 2000. Subtyping of uncultured bartonellae using sequence comparison of 16S/23S rRNA intergenic spacer regions amplified directly from infected blood. Mol. Cell. Probes 14:79-87. [DOI] [PubMed] [Google Scholar]
- 3.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891-897. [DOI] [PubMed] [Google Scholar]
- 4.Birtles, R. J., N. A. Saunders, T. G. Harrison, and D. H. Molyneux. 1995. Proposals to unify the genera Bartonella and Grahamella with descriptions of Bartonella talpae comb. nov., Bartonella peromyscii comb. nov., and three newly described species: Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int. J. Syst. Bacteriol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlin, J., L. Laughlin, S. Gordon, S Romero, N. Solorzano, and R. L. Regnery. 2000. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: test development and application to a population in an area of bartonellosis endemicity. J. Clin. Microbiol. 38:4269-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checkley, W., L. D. Epstein, R. H. Gilman, D. Figueroa, R. I. Cama, J. A. Patz, and R. E. Black. 2000. Effects of El Niño and ambient temperature on hospital admissions for diarrhoeal diseases in Peruvian children. Lancet 355:442-450. [DOI] [PubMed] [Google Scholar]
- 7.East, A. K., D. Allaway, and M. D. Collins. 1992. Analysis of DNA encoding 23S rRNA and 16S-23S rRNA intergenic spacer regions from Plesiomonas shigelloides. FEMS Microbiol. Lett. 95:57-62. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, B. A., L. D. Rotz, J. A. D. Leake, F. Samalvides, J. Bernable, G. Ventura, C. Padilla, P. Villaseca, L. Beati, R. Regnery, J. E. Childs, J. G. Olsen, and C. P. Carrillo. 1999. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am. J. Trop. Med. Hyg. 61:344-349. [DOI] [PubMed] [Google Scholar]
- 9.Fry, N. K., J. M. Bangsborg, S. Bernander, J. Etienne, B. Forsblom, V. Gaia, P. Hasenberger, D. Lindsay, A. Papoutsi, C. Pelaz, M. Struelens, S. A. Uldum, P. Visca, and T. G. Harrison. 2000. Assessment of intercentre reproducibility and epidemiological concordance of Legionella pneumophila serogroup 1 genotyping by amplified fragment length polymorphism analysis. Eur. J. Clin. Microbiol. Infect. Dis. 19:773-780. [DOI] [PubMed] [Google Scholar]
- 10.Gürther, V., and B. C. Mayall. 2001. Genomic approaches to typing, taxonomy and evolution of bacterial isolates. Int. J. Syst. E vol. Microbiol. 51:3-16. [DOI] [PubMed] [Google Scholar]
- 11.Hales, S., P. Weinstein, Y. Souares, and A. Woodward. 1999. El Niño and the dynamics of vector borne disease transmission. Environ. Health Perspect. 107:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handley, S. A., and R. L. Regnery. 2000. Differentiation of pathogenic Bartonella species by infrequent restriction site PCR. J. Clin. Microbiol. 38:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrer, A. 1990. Epidemiologia de la verruga peruana. Pribaceb, Lima, Peru.
- 14.Houpikian, P., P. E. Fournier, and D. Raoult. 2001. Phylogenetic position of Bartonella vinsonii subsp. arupensis based on 16S rDNA and gltA gene sequences. Int. J. Syst. E vol. Microbiol. 51:179-182. [DOI] [PubMed] [Google Scholar]
- 15.Kosek, M., R. Lavarello, R. H. Gilman, J. Delgado, C. Maguiña, M. Verastegui, A. G. Lescano, V. Mallqui, J. C. Kosek, S. Recavarren, and L. Cabrera. 2000. Natural history of infection with Bartonella bacilliformis in a non-endemic population. J. Infect. Dis. 182:865-872. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley and Sons, Chichester, United Kingdom.
- 17.Maguiña, C. 1995. Bartonellosis o enfermedad de Carrion. AFA Editores Importadores SA, Lima, Peru.
- 18.Minnick, M. F., J. C. Strange, and K. F. Williams. 1994. Characterization of the 16S-23S rRNA intergenic spacer of Bartonella bacilliformis. Gene 143:149-150. [DOI] [PubMed] [Google Scholar]
- 19.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux, V., and D. Raoult. 1995. The 16S-23S rRNA intergenic spacer region of Bartonella (Rochamilaea) species is longer than usually described for other bacteria. Gene 156:107-111. [DOI] [PubMed] [Google Scholar]
- 21.Sander, A., M. Ruess, S. Bereswill, M. Schuppler, and B. Steinbrueckner. 1998. Comparison of DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J. Clin. Microbiol. 36:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struelens, M. J., and members of the European Study Group on Epidemiological Markers (ESGEM) of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID). 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 23.Valsangiacomo, C., F. Baggi, V. Gaia, T. Balmelli, R. Peduzzi, and J.-C. Piffaretti. 1995. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J. Clin. Microbiol. 33:1716-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinman, D. 1944. Infectious anemias due to bartonella and related red cell parasites. Trans. Am. Phil. Soc. 33:243-287. [Google Scholar]
- 25.Weinman, D., and N. Pinkerton. 1937. Carrion's disease. IV. Natural sources of Bartonella in the endemic zone. Proc. Natl. Soc. Exp. Biol. Med. 37:596-598. [Google Scholar]
- 26.Young, L. J., and J. H. Young. 1998. Statistical ecology, a population perspective. Kluwer Academic Publishers, London, United Kingdom.