Abstract
The CRF01_AE (subtype E) strain of human immunodeficiency virus type 1 (HIV-1), originally reported in Thailand, spread rapidly to and showed prevalence in several countries in Southeast Asia, including Taiwan. This strain was also found in other regions of the world. Based on sequence analysis of the vpu gene, a nested PCR assay including an outer primer pair and a subtype E-specific inner primer pair was developed in this study for rapid detection of subtype E viruses. It was tested with 397 HIV-1-positive samples of known subtypes. For these samples, the sensitivity of detection of subtype E viruses was 100% (127 of 127), and the specificity was 97.8% (264 of 270). Although six samples of either subtype A or G showed a positive PCR, most of the cross-reactivity could be reduced by raising the annealing temperature from 54°C to 63°C. When tested with 195 HIV-positive samples of unknown subtypes, the assay had a sensitivity of 98.0% and a specificity of 98.6%. This is a simple, convenient, and sensitive method for rapid detection of subtype E viruses, especially in regions in which viruses of subtypes B and E are predominant.
Human immunodeficiency virus type 1 (HIV-1) displays an unusually high degree of genetic variability in vivo. Analyses of the env and gag genes have shown that HIV-1 can be classified into three groups: the M (major) group, the O (outlier) group, and the N (new) group (14, 24, 25, 27, 29, 38). Based on the env sequences, strains in the M group can be further divided into subtypes A through K (29, 41). The circulating recombinant form (CRF) of virus has been found in a significant proportion of HIV-1 infections (34). A recombinant strain, CRF01_AE, which was previously designated as subtype E and contained subtype E env and subtype A gag genes, was first reported in Thailand in 1993 (32). Subtype E virus spread rapidly and soon became the predominant HIV-1 type in Thailand. It also spread to and became prevalent in the neighboring countries in Southeast Asia (3, 4, 37), where more than two subtypes cocirculating in one area have frequently been reported. Subtype E was also prevalent on other continents (1, 2, 7, 28, 30). In Taiwan, subtype B and E viruses are the two most prevalent subtypes (23). The first subtype E-infected patient in Taiwan was recognized in 1989. Subtype E virus soon became prevalent in Taiwan. While subtype E virus now constituted about 20% of the HIV-1 infections in Taiwan, it accounted for a higher portion in the heterosexual transmission group (23).
Since the HIV epidemic is dynamic and the distribution of HIV-1 subtypes in a given population may change over time, it is necessary to periodically survey the circulating subtypes. Information on genetic subtypes is important for our understanding of the transmission and evolution of HIV-1; for study of viral pathogenesis, drug susceptibility, and viral load; and for global vaccine development (2, 8, 13, 33). It has been reported that subtype E virus is associated with a high risk of heterosexual transmission (19). To determine whether subtype E virus should be included in future vaccine development, detection of subtype E virus in a population is crucial.
Several techniques, such as DNA sequencing, heteroduplex mobility assay (HMA) (9), and hybridization with subtype-specific probes after PCR (11), have been utilized for identification of HIV-1 subtypes. In addition, serotyping with V3 peptide, which incorporates in the enzyme-linked immunosorbent assay (17), has also been successfully applied to studying the HIV-1 epidemic in Thailand (16). DNA sequencing requires sophisticated facilities and procedures and is therefore not practical as a routine survey method in developing countries. Although both HMA and DNA hybridization with subtype-specific probes after PCR are very sensitive and specific, they are time-consuming and laborious, especially when dealing with large numbers of samples (9, 40). Moreover, although the peptide-based assays work well for subtype E infection, with a specificity of 90.2% in one study, the feasibility of V3 serotyping in countries other than Thailand is still questionable (17, 40).
The HIV-1 vpu gene belongs to one of six accessory genes (26). It is located in the middle part of the genome, with its 3′ one-third overlapping the env gene. Previously, we demonstrated that the genetic subtypes determined by vpu sequence analysis correspond well to those determined by sequence analysis of the gag or env gene and that subtype E strains can be accurately determined by vpu gene analysis (23). In this study, we developed a simple method that incorporates subtype E-specific primers in the nested PCR of the vpu gene for detection of subtype E viruses. It can differentiate locally prevalent subtypes rapidly and elucidate the evolving molecular epidemiology.
The nucleotide sequences of the vpu gene of subtype B and subtype E HIV-1 strains, including 22 strains from GenBank and 92 Taiwanese strains sequenced in our laboratory, were aligned and compared first. Analysis of the consensus sequences of subtypes B and E revealed a nucleotide divergence of 20.3%. Within each subtype, while there was considerable sequence variation in the vpu genes among subtype B strains, the vpu genes among subtype E strains were rather conserved (data not shown). A PCR assay was therefore developed for rapid detection of subtype E strains. Two criteria were used in the primer design. First, the primers had to be located at the regions in which the two subtypes are most divergent. Second, the primers had to be located at the regions highly conserved among subtype E strains. Subtype E-specific primers VPUEN1 and VPUEN2 were thus designed. The sequences of VPUEN1 and VPUEN2 were 5′-TAGTGCAATAGTAGGACTGATAGTAGCG-3′ (genome positions 5650 to 5677 of the subtype E strain CM240) and 5′-CACAAGTTTGGCCAATTCATCTGT-3′ (genome positions 5813 to 5836), respectively.
Previously, we had designed two primer pairs, TAT-1/EN70 (outer pair) and 154.1/KPN (inner pair), which target the highly conserved regions upstream and downstream of the vpu gene, in a nested PCR assay to amplify a 398-bp product for both subtype B and E viruses (23). In this study, we adopted TAT-1 and EN70 as the outer primer pair and the subtype E-specific primers VPUEN1 and VPUEN2 as the inner primer pair in the nested PCR protocol. To test the feasibility of this assay for detection of subtype E viruses, we examined 397 HIV-1-positive blood samples of known subtypes collected at the Taipei Municipal Venereal Disease Center. Genomic DNA was isolated from peripheral blood mononuclear cells and quantitated as described previously (23, 31). One to 1.5 μg of genomic DNA was subjected to the first-round PCR with the outer primers (TAT-1 and EN70), and 1 μl of the first-round PCR product was subjected to the second-round PCR with the inner primers (154.1 and KPN) (23). Another microliter of the first-round PCR product was subjected to the subtype E PCR assay with the primers VPUEN1 and VPUEN2. The amplification conditions were 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 10 min. Standard precautions recommended for PCR were taken to avoid contamination (20).
One hundred twenty-seven subtype E and 259 subtype B samples, which were previously determined by nucleotide sequence analysis (23), were subjected to the subtype E PCR assay first. As exemplified in Fig. 1, strong bands of the expected size of 187 bp were seen in all of the subtype E samples, but in none of the subtype B samples. As the control, the conserved primers 154.1 and KPN were used in the second-round PCR, and the 398-bp PCR products were detected in all of these samples (Fig. 1). We next tested samples of subtypes A, C, and G by this method. Four out of five subtype G samples and two out of two subtype A samples gave positive results, although one of the subtype A samples showed a weak band (data not shown). All four subtype C samples were negative. These results are summarized in Table 1. For these samples of known subtypes, the sensitivity of detection of subtype E strains was 100%, and the specificity was 97.8% (264 of 270).
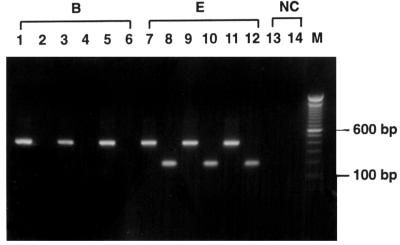
FIG. 1.
Detection of subtype E virus by the subtype E PCR assay. HIV-1 proviral DNA extracted from three samples each of subtypes B and E was subjected to the subtype E PCR assay (even-numbered lanes) and PCR with the conserved primers 154.1 and KPN (odd-numbered lanes). PCR products were electrophoresed through 1% agarose gel and stained with ethidium bromide. Products with the expected size of 187 bp were seen in the subtype E samples (lanes 8, 10, and 12). As the control, products of 398 bp containing the full-length vpu gene were amplified in all samples (odd-numbered lanes). A negative control (NC) was included in the experiment. Lane M, molecular size marker.
TABLE 1.
Reactivity of samples of known subtypes by the subtype E PCR assay
| Subtype | No. of samples with PCR result at annealing temp:
|
Total | |||
|---|---|---|---|---|---|
| 54°C
|
63°C
|
||||
| + | − | + | − | ||
| A | 2 | 0 | 1 | 1 | 2 |
| B | 0 | 259 | NDa | ND | 259 |
| C | 0 | 4 | 0 | 4 | 4 |
| E | 127 | 0 | 127 | 0 | 127 |
| G | 4 | 1 | 0 | 5 | 5 |
ND, not done.
The nucleotide sequences corresponding to the regions of VPUEN1 and VPUEN2 for these subtype A, C, and G strains were then aligned (Table 2). Compared with the VPUEN1 and VPUEN2 sequences, the subtype C strains differ by at least 18 nucleotides, and the subtype G strains differ by 7 to 10 nucleotides. One of the subtype A strains differs at 12 positions, and the other differs at 8 positions (Table 2).
TABLE 2.
Comparison of the nucleotide sequences in the vpu gene corresponding to the region of subtype E-specific primers
| Sample | Nucleotide sequence corresponding toa:
|
PCR resultb | |
|---|---|---|---|
| VPUEN1 | VPUEN2 | ||
| Consensus E | TAG.TGCAAT.AGTAGGACTGATAGTAGCG | ACAGATGAATTGGCCAAACTTGTG | |
| A1 | CT-.------.------------------- | -----G------T-A-C----A-T | + |
| A2 | ---G-TT--C.-A--CT-AGC--------A | --------------A--------- | + |
| C1 | ---.ATT-TAG-T-----G-AGG--CTCT- | -----G----------C-A-G--- | − |
| C2 | ---.ATT-TAG-T-----A-AGG--CCTT- | --T--G-----AT-A-C-A-G--- | − |
| C3 | ---.ATT-TAA-T-----G-AGG--C-TT- | --T--G-----AT-A-C-A-G--- | − |
| C4 | ---.ATT-TAG-T-----G-AGG--C-CT- | -----G----------C-A-G--- | − |
| G1 | ---C------.---------AG-------A | -----A-----AT---CT---A-- | + |
| G2 | ---C------.---------A--------A | -----G--------A-CC------ | + |
| G3 | ---C------.---------A--------A | -----G--------A-CC------ | + |
| G4 | ---C------.---------A--------A | -----G--------A-CC------ | + |
| G5 | ---C------.---------A--------A | -----G--------A-CC------ | + |
A dash indicates that the nucleotide is the same as the consensus subtype E sequence; a dot represents a space (no nucleotide at that position).
Subtype E-specific PCR at annealing temperature of 54°C.
To test whether the cross-reactivity of subtype A and G strains could be reduced by raising the annealing temperature in our subtype E PCR assay, the annealing temperature was increased (in increments of 2 to 3°C) empirically. It was found that an annealing temperature of 63°C could greatly reduce the cross-reactivity, while still preserving the reactivity of subtype E strains (data not shown). The results are summarized in Table 1. All 127 subtype E strains showed positive results, and only 1 subtype A strain (A1) was still reactive at an annealing temperature of 63°C.
To examine the applicability of this assay for detection of subtype E viruses from samples of unknown subtypes, 195 blood samples collected from the HIV-1-infected patients at the National Taiwan University Hospital were tested by this subtype E PCR assay in one laboratory. Fifty of these samples were positive, and 145 were negative (Table 3). DNA sequencing of the vpu genes of all these samples was independently performed in another laboratory with an ABI-373A autosequencer (Applied Biosystems, Foster City, Calif.) (23). Sequence analysis by a previously described method revealed that 48 of the 50 positive samples were subtype E and 2 were subtype G (18, 23). Of the 145 negative samples, 142 were subtype B, 1 was subtype E, 1 was subtype F, and 1 was subtype G. Application of this rapid assay to detect subtype E strains in these unknown samples revealed that the sensitivity was 98.0% and the specificity was 98.6% (144 of 146). Only one subtype E strain gave a negative result; however, it was positive in the repeated PCR experiment.
TABLE 3.
Correlation of the results obtained with the subtype E PCR assay and the subtypes determined by nucleotide sequence analysis
| Subtype determined by sequence analysis | No. of samples with subtype E-specific PCR resulta:
|
Total | |
|---|---|---|---|
| Positive | Negative | ||
| B | 0 | 142 | 142 |
| E | 48 | 1 | 49 |
| F | 0 | 1 | 1 |
| G | 2 | 1 | 3 |
| Total | 50 | 145 | 195 |
The annealing temperature for the PCR was 54°C.
Subtype E virus has been reported to be associated with a higher risk of heterosexual transmission than subtype B virus (19). Soto-Ramirez et al. demonstrated that subtype E virus grew more efficiently in Langerhans cells than subtype B virus, suggesting that Langerhans cell tropism is associated with the efficiency of heterosexual transmission of HIV-1 (39), although other studies did not support this hypothesis (10, 35). A recent study reported that the levels of plasma HIV RNA at the earliest time within 3 months of seroconversion were more than three times higher for persons infected with subtype E virus than for those infected with subtype B virus and suggested that initial viral load may be related to the biological difference between these two viruses (15). Whatever the mechanism, the observations that subtype E virus spread rapidly and became the predominant virus have been documented in many countries, especially those in Southeast Asia. In Thailand, approximately 96% of HIV-1-infected Thais carried subtype E virus (36). Subtype E infection also constituted a significant portion of the heterosexually transmitted group in Taiwan (23). Heterosexual men, mostly businessmen traveling back and forth, contracted subtype E virus in Thailand and brought it to Taiwan in the early 1990s.
In this study, a vpu gene-based PCR assay was developed for rapid detection of subtype E strains. Vpu has an amphipathic nature and consists of a hydrophobic N-terminal membrane anchor and a polar C-terminal cytoplasmic domain (12). A highly conserved region in the cytoplasmic domain has been shown to be critical for the biological function of Vpu (6). We have analyzed 114 vpu sequences, including 62 subtype B strains and 52 subtype E strains, to study the frequency of nucleotide substitution at each position. While comparison of the full-length vpu genes of the subtype E consensus and the subtype B consensus revealed a diversity of 20.3%, a higher degree of variation was found in the regions between nucleotides 18 to 45 and 181 to 204 of the vpu gene. Furthermore, the sequences in these two regions are relatively conserved in the subtype E viruses. We therefore designed the subtype E-specific primers on the basis of the sequences of these two regions in our PCR assay. When applied to 397 samples of known subtypes, this assay could successfully differentiate subtype E strains from subtype B strains.
The subtype E vpu PCR assay was also tested with 195 clinical samples of unknown subtypes. The samples with positive results were considered as subtype E, and those with negative results were considered as subtype B. Compared with the subtypes determined by vpu sequence analysis, the accuracy of the assay was 97.4% (190 of 195) (Table 3). Taking together the results from a total of 592 samples, all but 1 of the 176 subtype E samples could be detected. The one subtype E sample that gave a negative result was positive after the assay had been repeated. This could have been due to inadequate treatment of the sample or a technical error. On the other hand, the specificity was found to be 98.1% (408 of 416). Since the high specificity was based on samples of predominant subtypes B and E, an additional seven non-B/non-E samples, including three subtype C and four subtype G, were also examined by this assay. All three subtype C samples were negative. While the four subtype G samples were positive, they became negative after the annealing temperature had been raised to 63°C (data not shown). The specificity remained high (97.2% [411 of 423]).
Similar to other Southeast Asian countries, subtypes A, C, and F have also been found in Taiwan occasionally (5, 21, 23). In addition, subtype G has been identified in a few patients (22). The application of the vpu PCR assay in a population infected with multiple HIV subtypes needs to be addressed. Based on the vpu sequence analyses, subtypes A and G seem to have less nucleotide variation in the regions of so-called subtype E-specific primers (Table 2). Consistent with this, subtype A and G samples were positive in the subtype E PCR assay at an annealing temperature of 54°C. However, when the annealing temperature was raised to 63°C, most of the cross-reactivity could be reduced (Table 1). These findings suggest that further optimization of the PCR conditions can increase the specificity of this assay. Moreover, by the same approach, it is possible to design primers specific for other subtypes and develop subtype-specific PCR assays for rapid detection of other subtypes, especially in regions in which multiple HIV-1 subtypes cocirculate.
Subtypes B and E are the predominant subtypes in Taiwan and in many Southeast Asian countries as well (5, 23). The subtype E PCR assay developed in this study could rapidly and quite accurately differentiate subtype E from subtype B strains with high sensitivity and specificity. This assay is easy to perform and inexpensive. The method could be applied in large-scale studies. It would be especially useful in regions in which viruses of subtypes B and E are the predominant subtypes.
Acknowledgments
We are grateful to the staff of the Taipei Municipal Venereal Disease Control Institute for collection of samples and Chin-Der Lee, Wen-Sheng Fan, and Yi-Hong Lu for technical assistance.
This work was supported by a grant from the National Taiwan University Hospital (grant NTUH-89S1021) and grants from the Department of Health (grants DOH89-DC-1034 and DOH90-DC-1058) of Taiwan, Republic of China.
REFERENCES
- 1.Boni, J., H. Pyra, M. Gebhardt, L. Perrin, P. Burgisser, L. Matter, W. Fierz, P. Erb, J. C. Piffaretti, E. Minder, P. Grob, J. J. Burckhardt, M. Zwahlen, and J. Schupbach. 1999. High frequency of non-B subtypes in newly diagnosed HIV-1 infections in Switzerland. J. Acquir. Immune Defic. Syndr. 22:174-179. [DOI] [PubMed] [Google Scholar]
- 2.Brodine, S. K., R. A. Shaffer, M. J. Starkey, S. A. Tasker, J. L. Gilcrest, M. K. Louder, A. Barile, T. C. VanCott, M. T. Vahey, F. E. McCutchan, D. L. Birx, D. D. Richman, and J. R. Mascola. 1999. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann. Intern. Med. 131:502-506. [DOI] [PubMed] [Google Scholar]
- 3.Brown, T. M., K. E. Robbins, M. Sinniah, T. S. Saraswathy, V. Lee, L. S. Hooi, B. Vijayamalar, C. C. Luo, C. Y. Ou, J. Rapier, G. Schochetman, and M. L. Kalish. 1996. HIV type 1 subtypes in Malaysia include B, C and E. AIDS Res. Hum. Retrovir. 12:1655-1657. [DOI] [PubMed] [Google Scholar]
- 4.Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, M. O'Shaughnessy, and C. Y. Ou. 1996. Detection of HIV type 1 env subtypes A, B, C and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. [DOI] [PubMed] [Google Scholar]
- 5.Chang, K. S., C. I. Lin, M. O. Salminen, S. K. Liao, A. M. Wu, H. C. Lin, R. Y. Lin, and S. C. Twu. 1997. Diversity and distribution of gag and env subtypes among 146 HIV type 1 isolates in Taiwan. AIDS Res. Hum. Retrovir. 13:1539-1543. [DOI] [PubMed] [Google Scholar]
- 6.Chen, M.-Y., F. Maldarelli, M. K. Karczewski, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J. Virol. 67:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couturier, E., F. Damond, P. Roques, H. Fleury, F. Barin, J. B. Brunet, F. Brun-Vezinet, and F. Simon. 2000. HIV-1 diversity in France, 1996-1998. The AC 11 Laboratory Network. AIDS 14:289-296. [DOI] [PubMed] [Google Scholar]
- 8.Debyser, Z., E. Van Wijngaerden, K. Van Laethem, K. Beuselinck, M. Reynders, E. De Clercq, J. Desmyter, and A. M. Vandamme. 1998. Failure to quantify viral load with two of the three commercial methods in a pregnant woman harboring an HIV type 1 subtype G strain. AIDS Res. Hum. Retrovir. 14:453-459. [DOI] [PubMed] [Google Scholar]
- 9.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 10.Dittmar, M. T., G. Simmons, S. Hibbitts, M. O'Hare, S. Louisirirotchanakul, S. Beddows, J. Weber, P. R. Clapham, and R. A. Weiss. 1997. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J. Virol. 71:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelbrecht, S., and E. J. Rensburg. 1995. Detection of Southern African human immunodeficiency virus type 1 subtypes by polymerase chain reaction: evaluation of different primer pairs and conditions. J. Virol. Methods 55:391-400. [DOI] [PubMed] [Google Scholar]
- 12.Federau, T., U. Schubert, J. Flobdorf, P. Henklein, D. Schomburg, and V. Wray. 1995. Solution structure of the cytoplasmic domain of the human immunodeficiency virus type 1 encoded virus protein U (Vpu). Int. J. Pept. Protein Res. 47:297-310. [DOI] [PubMed] [Google Scholar]
- 13.Hammond, J., B. A. Larder, R. F. Schinazi, and J. W. Mellors. 1997. Mutations in retroviral gene associated with drug resistance, p. III-207-249. In Theoretical Biology and Biophysics Group (ed.), Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 14.Hu, D. J., T. J. Dondero, M. A. Rayfield, J. R. George, G. Schochetman, H. W. Jaffe, C. C. Luo, M. L. Kalish, B. G. Weniger, C. P. Pau, C. A. Schable, and J. W. Curran. 1996. The emerging genetic diversity of HIV: the importance of global surveillance for diagnostics, research, and prevention. JAMA 275:210-216. [PubMed] [Google Scholar]
- 15.Hu, D. J., S. Vanichseni, T. D. Mastro, S. Raktham, N. L. Young, P. A. Mock, S. Subbarao, B. S. Parekh, L. Srisuwanvilai, R. Sutthent, C. Wasi, W. Heneine, and K. Choopanya. 2001. Viral load differences in early infection with two HIV-1 subtypes. AIDS 15:683-691. [DOI] [PubMed] [Google Scholar]
- 16.Kalish, M. L., A. Baldwin, S. Raktham, C. Wasi, C. C. Luo, G. Schochetman, T. D. Mastro, N. Young, S. Vanichseni, H. Rubsamen-Waigmann, H. von Briesen, J. I. Mullins, E. Delwart, B. Herring, J. Esparza, W. L. Heyward, and S. Osmanov. 1995. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS 9:851-856. [DOI] [PubMed] [Google Scholar]
- 17.Kimdar, S., S. Anders, S. Juris, and S. Matti. 1994. Rapid grouping of a HIV-1 infection in subtypes A to E by V3 peptide serotyping and its relation to sequence analysis. Biochem. Biophys. Res. Commun. 205:1658-1664. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetic analysis, version 1.01. The Pennsylvania State University, University Park, Pa.
- 19.Kunanusont, C., H. M. Foy, J. K. Kreiss, S. Rerks-Ngarm, P. Phanuphak, S. Raktham, C. P. Pau, and N. L. Young. 1995. HIV-1 subtypes and male-to-female transmission in Thailand. Lancet 345:1078-1083. [DOI] [PubMed] [Google Scholar]
- 20.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. N., M. Y. Chen, H. S. Lin, M. C. Lee, C. C. Luo, S. J. Twu, R. Y. Lin, and C. Y. Chuang. 1998. HIV type 1 env subtype A variants in Taiwan. AIDS Res. Hum. Retrovir. 9:807-809. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C. N., M. Y. Chen, W. S. Fan, S. J. Twu, and R. Y. Lin. 1999. Domestic transmission of HIV type 1 subtype G strains in Taiwan. AIDS Res. Hum. Retrovir. 15:1137-1140. [DOI] [PubMed] [Google Scholar]
- 23.Lee, C.-N., W.-K. Wang, W.-S. Fan, S.-J. Twu, S.-C. Chen, M.-C. Sheng, and M.-Y. Chen. 2000. Determination of human immunodeficiency virus type 1 subtypes in Taiwan by vpu gene analysis. J. Clin. Microbiol. 38:2468-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitner, T. 1996. Genetic subtypes of HIV-1, p. III-28-40. In Theoretical Biology and Biophysics Group (ed.), Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 25.Louwagie, J., F. E. McCutchan, M. Peeters, T. P. Brennan, E. Sanders-Buell, G. A. Eddy, G. van der Groen, K. Fransen, G.-M. Gershy-Damet, R. Deleys, and D. S. Burke. 1993. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS 7:769-780. [DOI] [PubMed] [Google Scholar]
- 26.Luciw, P. A. 1996. Human immunodeficiency viruses and their replication, p. 1881-1952. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.McCutchan, F. E., M. O. Salminen, J. K. Carr, and D. S. Burke. 1996. HIV-1 genetic diversity. AIDS 10(Suppl. 3):S13-S20. [PubMed] [Google Scholar]
- 28.Murphy, E., B. Korber, M. C. Georges-Courbot, B. You, A. Pinter, D. Cook, M. P. Kieny, A. Georges, C. Mathiot, F. Barre-Sinoussi, and M. Girard. 1993. Diversity of V3 region sequences of human immunodeficiency viruses type 1 from the Central African Republic. AIDS Res. Hum. Retrovir. 9:997-1006. [DOI] [PubMed] [Google Scholar]
- 29.Myers, G. 1993. Assimilating HIV sequences. AIDS Res. Hum. Retrovir. 9:697-702. [DOI] [PubMed] [Google Scholar]
- 30.Op De Coul, E. L., R. A. Coutinho, A. van Der Schoot, G. J. van Doornum, V. V. Lukashov, J. Goudsmit, and M. Cornelissen. 2001. The impact of immigration on env HIV-1 subtype distribution among heterosexuals in The Netherlands: influx of subtype B and non-B strains. AIDS 15:2277-2286. [DOI] [PubMed] [Google Scholar]
- 31.Ou, C. Y., S. Kwok, S. W. Mitchell, D. H. Mack, J. J. Sninsky, J. W. Krebs, P. Feorino, D. Warfield, and G. Schochetman. 1988. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science 239:295-297. [DOI] [PubMed] [Google Scholar]
- 32.Ou, C. Y., Y. Takebe, B. G. Weniger, C. C. Luo, M. L. Kalish, W. Auwanit, S. Yamazaki, H. D. Gayle, N. L. Young, and G. Schochetman. 1993. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet 341:1171-1174. [DOI] [PubMed] [Google Scholar]
- 33.Palmer, S., A. Alaeus, J. Albert, and S. Cox. 1998. Drug susceptibility of subtypes A, B, C, D, and E human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 14:157-162. [DOI] [PubMed] [Google Scholar]
- 34.Peeters, M. 2000. Recombinant HIV sequences: their role in the global epidemic, p. I-39-54. In Theoretical Biology and Biophysics Group (ed.), HIV sequence compendium 2000. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 35.Pope, M., S. S. Frankel, J. R. Mascola, A. Trkola, F. Isdell, D. L. Birx, D. S. Burke, D. D. Ho, and J. P. Moore. 1997. Human immunodeficiency virus type 1 strains of subtypes B and E replicate in cutaneous dendritic cell-T-cell mixtures without displaying subtype-specific tropism. J. Virol. 71:8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruxrungtham, K., and P. Phanuphak. 2001. Update on HIV/AIDS in Thailand. J. Med. Assoc. Thai. 84(Suppl. 1):S1-S17. [PubMed] [Google Scholar]
- 37.Santiago, M. L., E. G. Santiago, J. C. Hafalla, M. A. Manalo, L. Orantia, M. N. Cajimat, C. Martin, C. Cuaresma, C. E. Dominguez, M. E. Borromeo, A. S. De Groot, T. P. Flanigan, C. C. Carpenter, K. H. Mayer, and B. L. Ramirez. 1998. Molecular epidemiology of HIV-1 infection in the Philippines, 1985 to 1997: transmission of subtypes B and E and potential emergence of subtypes C and F. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 18:260-269. [DOI] [PubMed] [Google Scholar]
- 38.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 39.Soto-Ramirez, L. E., B. Renjifo, M. F. McLane, R. Marlink, C. O'Hara, R. Sutthent, C. Wasi, P. Vithayasai, V. Vithayasai, C. Apichartpiyakul, P. Auewarakul, V. Pena Cruz, D. S. Chui, R. Osathanondh, K. Mayer, T. H. Lee, and M. Essex. 1996. HIV-1 Langerhans' cell tropism associated with heterosexual transmission of HIV. Science 271:1291-1293. [DOI] [PubMed] [Google Scholar]
- 40.Subbarao, S., K. Limpakarnjanarat, T. D. Mastro, J. Bhumisawasdi, P. Warachit, C. Jayavasu, N. L. Yang, C. C. Luo, N. Shaffer, M. L. Kalish, and G. Schochetman. 1998. HIV type 1 in Thailand, 1994-1995: persistence of two subtypes with low genetic diversity. AIDS Res. Hum. Retrovir. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 41.Triques, K., A. Bourgeois, N. Vidal, E. Mpoudi-Ngole, C. Mulanga-Kabeya, N. Nzilambi, N. Torimiro, E. Saman, E. Delaporte, and M. Peeters. 2000. Near-full-length genome sequencing of divergent African HIV type 1 subtype F viruses leads to the identification of a new HIV type 1 subtype designated K. AIDS Res. Hum. Retrovir. 16:139-151. [DOI] [PubMed] [Google Scholar]