Abstract
Sensitive and early detection of emerging hepatitis B virus (HBV) drug resistance may not only help monitor the viral dynamics associated with lamivudine treatment but could also improve therapeutic decision making. This is especially important when new antivirals effective against lamivudine-resistant HBV become available. A total of 159 serum samples from 33 chronic HBV patients receiving lamivudine treatment were analyzed at four centers for the presence of lamivudine-resistant mutations at codons 528 [180] (proposed revised nomenclature according to Stuyver et al. [Hepatology 33:751-757, 2001] shown in brackets), 552 [204], and 555 [207] of the HBV polymerase. Sequencing data were compared with results generated by the INNO-LiPA HBV DR line probe assay (LiPA), an assay based on reverse hybridization of amplified HBV DNA fragments with specific nucleotide probes immobilized on nitrocellulose strips. LiPA provided at least the same information as sequencing for 97.5% of all codons analyzed for codon 528 [180], 95% for codon 552 [204], and 100% for codon 555 [207]. The most common reason for discrepant or indeterminate results (0.4% and 1.5%, respectively) in a small percentage of the population tested could be attributed to polymorphisms not yet covered by LiPA probes. In at least five patients, a mutant could be detected earlier by LiPA than by sequencing. In 15 patients, LiPA detected mixed wild-type and mutant virus populations before viral breakthrough. These results demonstrate that INNO-LiPA HBV DR is a highly sensitive and easily applicable assay for the detection and monitoring of lamivudine-resistant mutations in chronic hepatitis B patients and that the assay is more sensitive than sequencing in detecting mixed mutant and wild-type sequences.
Although safe and effective vaccines for the prevention of hepatitis B virus (HBV) infection have been available for almost 20 years, the disease remains a major cause of morbidity and mortality worldwide. It is currently estimated that 350 million people are chronic carriers of the virus, often as a result of infection during childhood. Approximately one third to one quarter of these individuals will develop progressive liver disease, including cirrhosis and primary hepatocellular carcinoma (13, 15). Some 1.2 million people die prematurely each year from conditions directly related to HBV infection.
Current treatment of chronic hepatitis B aims at interrupting the progression and clinical outcomes of the disease by suppressing viral replication, as evidenced by hepatitis B virus E antigen seroconversion to hepatitis B virus E antibodies (14) or by a decrease in viral load. The first approved therapeutic agent was alpha interferon. Unfortunately, alpha interferon is expensive, is effective in no more than 30% of patients, has to be administered by injection, and is associated with numerous side effects which may necessitate dosage reduction or even treatment discontinuation (13, 15).
In 1998, the nucleoside analogue lamivudine was approved for use in patients with chronic hepatitis B (7). The convenience of a one-pill-per-day regimen and the low incidence of side effects make it a preferred treatment for many physicians and patients. However, viral breakthrough is detected in approximately 16% to 32% of patients after 1 year of treatment (12). Newer oral nucleoside/nucleotide analogues in clinical trials, such as adefovir dipivoxil (4), entecavir (6), and emtricitabine (19, 18), appear to be at least as potent as lamivudine. In vitro and in vivo studies showed that adefovir and entecavir are also effective in suppressing lamivudine-resistant HBV (8).
Mutations in codon 552 [204] (proposed revised nomenclature according to Stuyver et al. [22] is shown in brackets) within the YMDD (tyrosine-methionine-aspartic acid-aspartic acid) motif of the HBV reverse transcriptase/polymerase with substitution of the methionine for valine or isoleucine (M552V/I [M204V/I]) are implicated in the decrease of viral susceptibility to lamivudine (1, 5, 10, 11). In addition, mutations in codons 528 [180] (2, 3, 11) and 555 [207] (9, 17) have also been linked to lamivudine and famciclovir resistance. Sensitive and early detection of emerging resistance may not only help monitor the viral dynamics associated with treatment but could also improve therapeutic decision making, especially as new antivirals that are effective against lamivudine-resistant HBV become available.
The INNO-LiPA HBV DR line probe assay (LiPA) is the first commercial assay designed to detect the presence of different genetic variants of HBV containing mutations located at amino acid positions 528 [180], 552 [204], and 555 [207] in the HBV polymerase protein, which confer resistance to lamivudine (21). The assay provides a rapid and easy-to-use alternative to sequence analysis for the detection of lamivudine-resistant mutations. We compared the results of this assay with those of direct sequencing for concordance in a geographically diverse population of HBV-infected patients who had developed resistance to lamivudine treatment. In addition, we sought to determine whether the emergence of lamivudine-resistant mutants could be detected earlier with INNO-LiPA HBV DR than with sequencing, as LiPA reverse hybridization technology seems to be well suited for the detection of mixed wild-type and mutant viral populations (1, 16, 24).
MATERIALS AND METHODS
Sample collection.
Samples from 33 hepatitis B patients who developed breakthrough infection during lamivudine treatment, defined by rebound of viral load or the presence of a lamivudine-resistant mutant on sequencing, were analyzed in four different centers: the Victorian Infectious Diseases Reference Laboratory, North Melbourne, Australia (center 1); INSERM Unit 271, Lyon, France (center 2); University of Michigan Medical Center, Ann Arbor, Mich. (center 3); and Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy (center 4). A serum sample of 0.5 to 1 ml was requested for testing and possible further investigation. As LiPA results were to be compared with those of sequencing, HBV polymerase open reading frame sequence data were requested for all samples.
The following information was recorded for each sample: date of sampling, HBV DNA level at the time of sampling, method used for viral load quantification, and the sequencing result at the time of sampling. Optionally, alanine aminotransferase levels and the presence of hepatitis B virus E antigen and anti-hepatitis B virus E antibodies were recorded. In addition, the start date of lamivudine treatment, indication for treatment (recurrent hepatitis B posttransplant or chronic hepatitis B), previous treatment (Table 1), and hepatitis C virus or human immunodeficiency virus coinfection were noted.
TABLE 1.
Previous treatmentsa
| Previous treatment | No. of patients |
|---|---|
| No information | 5b |
| None | 8 |
| IFN-α | 11c |
| IFN-α + Fam | 3 |
| IFN-α + other | 3 |
| IFN-α + Fam + other | 2 |
| Ganciclovir | 1 |
Treatment received before initiation of lamivudine treatment. IFN-α, alpha interferon; Fam, famciclovir, other, ganciclovir, vidarabine, or ribavirin.
One patient ceased lamivudine treatment and was further treated with adefovir dipivoxil.
One patient continued alpha interferon treatment during lamivudine treatment.
INNO-LiPA HBV DR.
The principle of the INNO-LiPA HBV DR assay has been described previously (23). The test was performed according to the manufacturer's instructions (Innogenetics N.V., Ghent, Belgium). Briefly, DNA was isolated from the serum samples. Then 10 μl of the extracted DNA was used for the first-round and 2 μl of the amplified product was used for the second-round PCR amplification. PCR conditions were as described in the product insert. Both PCR products were visualized on a 2% agarose gel. Usually the second-round amplification product was used for hybridization to the LiPA strips as described in the product insert, except in one center, where the first-round product was used when visible on an agarose gel. All centers first tested a proficiency panel to demonstrate adequate experience with the technique.
The probes on the INNO-LiPA HBV DR strip cover the following amino acids: codon 528 [180], wild-type leucine (L) and mutant methionine (M); codon 552 [204], wild-type methionine and mutants valine (V) and isoleucine (I); codon 555 [207], wild-type valine, leucine, and methionine and mutant isoleucine. The assay cannot detect any mutations other than those on the strip. The nomenclature according to genotype A was used here for practical reasons, in that the same nomenclature was used in the INNO-LiPA HBV DR kits. An alternative nomenclature, generally applicable for all genotypes, has been proposed recently by Stuyver et al. (22), in which codons 528, 552, and 555 correspond to codons 180, 204, and 207, respectively. The alternative numbering is shown in brackets.
Sequencing.
The sequencing method was not standardized across the centers, with samples sequenced at each center by their own in-house method. When no sequence data were available and sufficient sample was present, the sequences were generated at Innogenetics. Wild-type, mutation, or mixed status was scored for codons 528 [180], 552 [204], and 555 [207].
Viral load.
The viral load of the samples was determined at the centers with a routinely used, commercially available quantitative assay from Digene Corporation, Gaithersburg, Md. (centers 1 and 4), Chiron Corporation, Emeryville, Calif. (center 2), or Abbott Laboratories, Abbott Park, Ill. (center 3), according to the manufacturer's instructions.
Clonal analysis of viral populations.
For confirmation of the presence of minor virus populations in a sample, PCR fragments were generated with nonbiotinylated versions of the PCR primers present with the INNO-LiPA HBV DR kit. These PCR fragments were subsequently cloned in the pGEM-T vector (Promega, Madison, Wis.), and inserts were sequenced with vector-specific primers. Five clones were initially sequenced for each sample; additional clones were sequenced when the results proved inconclusive (i.e., when the presence of a mixed population could not be demonstrated).
RESULTS
Patients and samples.
A total of 159 serum samples from 33 patients were analyzed, ranging from 3 to 16 samples per patient sampled over a 9- to 47-month period. Thirty patients were male, and three were female. The mean age was 49.7 years (range, 27 to 65 years). Twenty-seven patients were Caucasian, 3 were Oriental, 2 were African, and 1 was Polynesian. Ten had recurrent hepatitis B post-liver transplantation; the other patients had histologically proven chronic hepatitis B. One patient was coinfected with hepatitis C virus. None of the patients were coinfected with human immunodeficiency virus or hepatitis D virus.
The spread in geographic location (North America, Australia, and Europe) of the study centers ensured that the HBV-positive samples studied represented different genotypes. Genotype D was the predominant genotype (22 of 33), and genotypes A (5 of 33) and C (5 of 33) were equally prevalent. One patient was infected with genotype B. Caucasians were predominantly genotype D (20 of 27) or A (5 of 27), and patients of Oriental origin were either genotype B (1 of 3) or C (2 of 3). Both African patients were genotype D.
Eight of the 33 patients had received no previous drug treatment; the remainder had been treated before for HBV infection, mainly with alpha interferon and famciclovir. One patient was also receiving alpha interferon at the same time as lamivudine treatment (Table 1).
Distribution of mutations.
On the basis of the sequencing results, we observed that the M552V [M204V] mutation was found in 18 (54.5%) patients, 17 of whom also had the L528M [L180M] mutation. The M552I [M204I] mutation was observed in 10 (30.3%) patients, in four cases in combination with the L528M [L180M] mutation. In addition, one patient showed an M552I-to-M552V [M204I-to-M204V] switch. This patient also carried the L528M [L180M] mutation. Four patients had only wild-type virus as far as sequencing data were available. A mutation at codon 555 [207] or a 552 wild-type (552WT)/L528M [204WT/L180M] combination was never observed with sequencing.
Concordance between LiPA and sequencing.
Sequencing and LiPA results were available for comparison for 159 samples. More than 80% of all sequencing data were obtained from the centers; the others were generated at Innogenetics. Results obtained for codon 528 [180], 552 [204], and 555 [207] by LiPA and by sequence analysis are summarized in Table 2. Full concordance between LiPA and sequence analysis was observed in 88.9% (424 of 477) of the codons analyzed. In a further 8.6% (41 of 477) of the codon results, LiPA provided additional information compared to sequencing. LiPA showed at least the same information as sequencing in 97.5% of the cases for codon 528 [180], in 95% of the cases for codon 552 [204], and in 100% of the cases for codon 555 [207]. For the last, the wild-type virus was observed with both techniques in all samples, except for one sample where a mix of wild-type and mutant viruses was found with LiPA, while sequencing detected only the wild type.
TABLE 2.
Comparison of sequence data and LiPA results: overview of results obtained at the four participating centers
| Codon | No. (%) of samples showing full concordance | No. of samples showing full concordance
|
No. of samples showing mix on LiPA but not sequencing (no. wild type/no. mutant) | No. of samples
|
Complete discordance | Total no. of codon results | |||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | Mutant | Mix | Mix by sequencing but not by LiPA | Indeterminate by LiPA | |||||
| 528 [180] | 130 (81.8) | 95 | 33 | 2 | 25 (3/22) | 3a | 1 | 159 | |
| 552 [204] | 136 (85.5) | 72 | 62 | 2 | 15 (5/10) | 2 | 6 | 159 | |
| 555 [207] | 158 (99.4) | 158 | 1 (1/0) | 159 | |||||
| Total | 424 (88.9) | 325 | 95 | 4 | 41 (9/32) | 3 | 2 | 7 | 477 |
Upon retesting at Innogenetics, LiPA results were concordant with sequencing results.
Samples showing a mixed virus population.
In 41 codon results (8.6% of codons analyzed), LiPA revealed the presence of both mutant and wild-type viruses, while sequencing detected only wild-type or mutant virus (Table 2). These results were of particular interest because they provided information about the ability of the LiPA to detect a mixed wild-type and mutant virus population earlier than sequencing or to detect a disappearing virus population for a longer period of time. For this reason, the nine codon results representing six patients in whom LiPA detected a mix of mutant and wild-type virus while sequencing detected only wild-type virus were analyzed in more detail.
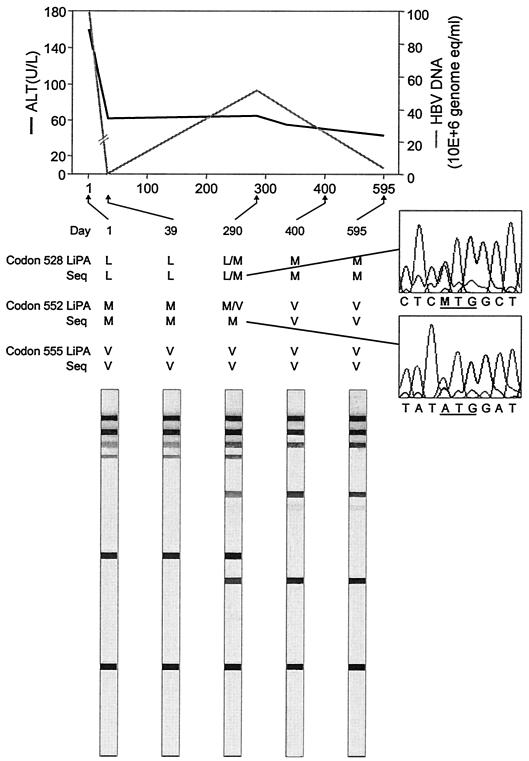
In five of these nine codon results, the mutant virus was detected by sequencing in samples taken at a later point in time, clearly indicating that the mutant was indeed already present in the previous sample and that LiPA detected this virus population before sequencing did. A representative example is shown in Fig. 1.
FIG. 1.
Viral load and alanine aminotransferase (ALT) response during lamivudine treatment. Serial samples from one patient were analyzed by LiPA and sequencing (Seq). Amino acids are indicated with the one-letter code: leucine (L), methionine (M), and valine (V). Days of follow-up are indicated above each strip.
For one codon result, LiPA showed a mix with a weak band for the mutant, while sequencing showed only the wild type. As the viral load was also increased, after having dropped following the start of treatment, it was decided to sequence this sample again. This second sequencing result indicated a clear mutant pattern, confirming the LiPA findings that both mutant and wild-type viruses were present.
In another patient, a mix of wild-type and I555 [I207] mutant viruses was observed in a sample taken 2 months after treatment. The mutant was not detected in the following samples and was only found again in the last follow-up sample taken 3 years later (as no sequencing data were available for this sample, it was not included in the final analysis). This result suggests that a minority population carrying the I555 [I207] mutation was present at a concentration that was close to the detection limit of the LiPA. Alternatively, the finding may be related to the intermittent presence of virus carrying this mutation (17).
In one case, the patient had ceased lamivudine treatment after emergence of treatment-resistant virus (M552I [M204I]). Only wild-type virus was detected by sequencing in the sample taken after treatment cessation, while in the previous two samples, the mutation M552I [M204I] was found. However, LiPA could still detect the mutant after treatment cessation (a mix of mutant and wild-type viruses was found with LiPA).
In another patient, one sample showing a mix on LiPA was collected 4 months before the start of lamivudine therapy, and the samples collected before and after that date were found to be wild type. The presence of the mutant has not been confirmed.
Indeterminate results.
In two patients (0.4% of codons analyzed), LiPA results for codon 552 [204] were indeterminate in that they were negative for both wild-type and mutant viruses (Table 2). In one case, detailed sequence analysis revealed that this result was due to a mutation near codon 552 [204], creating a polymorphism that interfered with the annealing of the LiPA probes. No polymorphism was observed in the second sample.
Completely discordant results.
For seven samples (1.5% of codons analyzed) from two patients, complete discordance between sequencing and LiPA results was observed for one codon (Table 2). Discrepancies were observed in codon 552 [204] in six samples from the same patient. Again, these results could be attributed to a mutation in a nearby codon creating a polymorphism that inhibited probe annealing. In these samples, minor virus populations without the polymorphism were still present and were detected by LiPA. Because the minor virus populations were not detected by sequence analysis, the two procedures yielded discrepant results.
Our hypothesis was confirmed by clonal analysis of two of the six samples, revealing a mixture of virus populations with and without the polymorphism. The clones carrying the polymorphism (and thus not detectable with LiPA) all had M552V [M204V], whereas the clones without the polymorphism all had M552I [M204I]. The other discrepant result was observed in codon 528 [180], where sequencing indicated the presence of a mutant while LiPA detected the wild type. Retesting of the LiPA result at Innogenetics indicated the presence of both mutant and wild-type viruses.
Detection of mutants by LiPA in relation to viral breakthrough.
In 15 (45.5%) patients, the mutant was detected by LiPA before viral breakthrough. In one representative patient, only wild-type virus was detected at the start of treatment. Twenty-nine weeks after treatment initiation, LiPA detected a mix of L528M/M552V [L180M/M204V] mutant and wild-type viruses, while viral load remained low. Viral breakthrough was diagnosed 17 weeks later. At that time, only the mutant virus was found. In the remaining 18 patients, mutant detection coincided with the increase in viral load.
DISCUSSION
The performance of the LiPA HBV DR assay for detection of mutations associated with lamivudine resistance was compared with that of direct sequencing for 33 chronically HBV-infected patients who had developed resistance to lamivudine treatment. The results show that the LiPA is at least as sensitive as sequencing in detecting treatment-resistant mutations and more sensitive in the early detection of mixed viral populations. LiPA may thus be able to detect a disappearing mutant population after treatment cessation for a longer period of time or to detect the emergence of a mutant earlier during antiviral therapy.
In this study, LiPA detected the mutant population earlier than sequencing in five of nine patients, where LiPA detected both mutant and wild-type viruses while sequence analysis found only the wild type. This was confirmed when both sequence analysis and LiPA subsequently detected the mutant virus in samples taken at a later date. LiPA also detected the persistence of mutant virus for at least 5 months longer than sequencing in one patient who had stopped treatment after emergence of the M552I [M204I] mutation. LiPA might therefore also be a valuable tool for monitoring patients in drug interruption programs. With numerous candidate antiviral agents for hepatitis B in late-stage clinical development, the availability of a sensitive and reliable drug resistance assay could assist in optimizing patient treatment.
The high sensitivity of LiPA observed in the present study has also been reported in other investigations. Aberle et al. (1) reported detection of a virus concentration as low as 103 copies/ml with LiPA and a higher sensitivity for detecting minority populations compared to sequencing. Another recent study (16) showed that a minority population comprising only 10% of the total viral load could be detected with LiPA, while the detection limit of sequence analysis was 50%. In addition, mutant virus could still be detected by LiPA for 3 to 46 weeks after cessation of lamivudine treatment, while earlier reports based on sequencing suggested that mutants disappear rapidly after treatment withdrawal (5). These findings therefore have relevant implications for future antiviral clinical studies, as mutant virus can still be present long after the 6-month wash-out period used in most clinical trials. They also indicate that patients with breakthrough infection secondary to lamivudine-resistant mutants are unlikely to respond to retreatment with lamivudine or other antiviral agents that are cross-resistant long after lamivudine is discontinued.
Clonal analysis showed that the most common reason for discrepant or indeterminate results could be attributed to polymorphisms not yet covered by LiPA probes. These included mutations at codons 529 [181] and 550 [202], prohibiting both wild-type and mutant LiPA probes from annealing at codons 528 [180] and 552 [204], respectively. These polymorphisms can give rise to a discordant result if a small population of mutant virus without the polymorphism is detected by LiPA and not by sequencing or yield an indeterminate result if the entire mutant virus population carries the polymorphism and is not detected by LiPA but is detected by sequencing.
It is quite possible that the different sequencing techniques used at each center can introduce variability when comparing the techniques. However, each center used a standard, sensitive sequencing technique, and all chromatograms were visually rechecked for the presence of mixtures. Moreover, a study of 48 Chinese HBV patients (E. Sablon, F. M. Yuen, C. L. Lai, H. Decraemer, and G. Maertens, Hepatology 34:321A, abstract 595, 2001), with the same amplicon used for direct sequencing of the PCR product and for INNO-LiPA HBV DR, also demonstrated superior sensitivity of the INNO-LiPA for detection of mixed wild-type and mutant populations.
The case in which the result of the repeat sequencing differed from the first test does suggest that the PCR preceding the sequence analysis can bias the final result, e.g., when two virus populations are present and the DNA of one population is preferentially amplified. PCR bias would normally be expected to occur equally in both assays. However, our results and the results of a second study (E. Sablon et al., abstract 595) showed that a mixture on sequencing but no mixture on LiPA is rarely found, while the opposite is observed more frequently. We therefore believe that these discrepancies can predominantly be attributed to the difference in sensitivity between the assays rather than PCR bias.
The distribution of mutations observed in this study is in agreement with other reports that L528M/M552V [L180M/M204V] is commonly found in patients resistant to lamivudine treatment. Contrary to initial reports (25, 26), L528M/M552I [L180M/M204I] was also observed, albeit to a lesser extent.
In about 50% of the patients with breakthrough infection, lamivudine-resistant mutations could be detected prior to viral breakthrough. This number is probably an underestimation due to the long intervals between sampling in some patients. These results are in agreement with the findings of Si Ahmed et al. (20), who reported detection of drug-resistant mutations with the LiPA several months before the occurrence of viral breakthrough.
The INNO-LiPA HBV DR assay allows simultaneous detection of wild-type sequences and key mutations at codons 528 [180], 552 [204], and 555 [207] of the HBV polymerase gene that have been reported to be associated with lamivudine resistance. The test is simple and allows rapid identification of treatment-resistant mutations in multiple patients with one assay. The INNO-LiPA is therefore well suited for use in a clinical setting to monitor hepatitis B patients receiving antiviral therapy. As newer antiviral agents become available, the ability of the LiPA assay to detect mutant virus populations earlier than sequencing and before viral breakthrough will permit timely initiation of new therapies effective against lamivudine-resistant HBV.
The test can also be used to determine the cross-resistance of lamivudine-resistant mutations to other antiviral agents. When mutations associated with resistance to new antiviral drugs are identified, the INNO-LiPA HBV DR can easily be updated to detect these new mutations. The LiPA can also help to monitor treatment compliance, as viral breakthrough in the absence of mutant virus might be indicative of nonadherence to the treatment protocol. In addition, the ability of the LiPA to detect mutant virus populations long after cessation of lamivudine treatment permits appropriate selection of antiviral agents for retreatment.
Acknowledgments
We gratefully acknowledge the assistance of J. Brissinck (Innogenetics N.V.) in writing and revising the manuscript. T. James and F. Shapiro (Innogenetics N.V.) are thanked as well for useful editorial comments. We also thank A. Barthelomeusz, M. Littlejohn, A. Ayres and L. Tracey (Victorian Infectious Diseases Reference Laboratory) as well as C. Pichoud (INSERM Unit 271) and M. Hussain (University of Michigan Medical Center) for valuable technical assistance in the study.
REFERENCES
- 1.Aberle, S. W., J. Kletzmayr, B. Watschnger, B. Schmied, N. Vetter, and E. Puchhammer-Stöckl. 2001. Comparison of sequence analysis and the INNO-LiPA HBV DR line probe assay for detection of lamivudine-resistant hepatitis B virus strains in patients under various clinical conditions. J. Clin. Microbiol. 39:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aye, T. T., A. I. Barthelomeusz, T. Shaw, A. Breshkin, L. Gronen, S. Bowden, J. McMillan, and S. Locarnini. 1996. Hepatitis B virus polymerase mutations during famciclovir therapy in patients following liver transplantation. Hepatology 24(Suppl.):285A. [DOI] [PubMed] [Google Scholar]
- 3.Aye, T. T., A. I. Barthelomeusz, T. Shaw, S. Bowden, A. Breshkin, J. McMillan, P. Angus, and S. Locarnini. 1997. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J. Hepatol. 26:1148-1153. [DOI] [PubMed] [Google Scholar]
- 4.Benhamou, Y., M. Bochet, V. Thibault, V. Calvez, M. H. Fievet, P. Vig, G. C. Gibbs, C. Brosgard, J. Fry, H. Namini, C. Katlama, and T. Poynard. 2001. Safety and efficacy of adefovir dipivoxil in patients co-infected with human immunodeficiency virus-1 and lamivudine-resistant hepatitis B virus: an open label study. Lancet 358:718-723. [DOI] [PubMed] [Google Scholar]
- 5.Chayama, K., Y. Suzuki, M. Kobayashi, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 6.De Man, R. A., L. Wolters, F. Nevens, D. Chua, M. Sherman, C. Lai, A. Gadano, Y. Lee, F. Mazzotta, N. Thomas, and D. De Hertogh. 2001. Safety and efficacy of oral entecavir given for 28 days in patients with chronic hepatitis B virus infection. Hepatology 34:578-582. [DOI] [PubMed] [Google Scholar]
- 7.Dienstag, J. L., E. R. Schiff, M. Mitchell, D. E. Casey, Jr., N. Gitlin, T. Lissoos, L. D. Gelb, L. Condreay, L. Crowther, M. Rubin, and N. Brown. 1999. Extended lamivudine retreatment for chronic hepatitis B: maintenance of viral suppression after discontinuation of therapy. Hepatology 30:1082-1087. [DOI] [PubMed] [Google Scholar]
- 8.Farrell, G. C. 2000. Clinical potential of emerging new agents in hepatitis B. Drugs 60:701-710. [DOI] [PubMed] [Google Scholar]
- 9.Günther, S., F. van Breunig, S. Santantonio, M.-C. Jung, G. B. Gaeta, L. Fisher, M. Sterneck, and H. Will. 1999. Absence of mutations in the YMDD motif/B region of the hepatitis B virus polymerase in famciclovir therapy failure. J. Hepatol. 30:749-754. [DOI] [PubMed] [Google Scholar]
- 10.Hunt, C., J. M. McGill, M. I. Allen, and L. D. Condreay. 2000. Clinical relevance of hepatitis B viral mutations. Hepatology 31:1037-1044. [DOI] [PubMed] [Google Scholar]
- 11.Hussain, M., and A. S. F. Lok. 1999. Mutations in the hepatitis B virus polymerase gene associated with antiviral treatment for hepatitis B. J. Viral Hepatol. 6:183-194. [DOI] [PubMed] [Google Scholar]
- 12.Lai, C. L., R. W. Chine, N. W. Y. Leung, T. T. Chang, R. Guan, D. I. Tai, K. Y. Ng, P. C. Wu, J. C. Dent, J. Barber, S. L. Stephenson, and D. F. Gray. 1998. A one-year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Lin, O. S., and E. B. Keeffe. 2001. Current treatment strategies for chronic hepatitis B and C. Annu. Rev. Med. 52:29-49. [DOI] [PubMed] [Google Scholar]
- 14.Lueng, N. 2000. Nucleoside analogues in the treatment of chronic hepatitis B. J. Gastroenterol. Hepatol. 15(Suppl.):E53-E60. [DOI] [PubMed] [Google Scholar]
- 15.Maddrey, W. C. 2001. Hepatitis B—an important public health issue. Clin. Lab. 47:51-55. [PubMed] [Google Scholar]
- 16.Pas, S. D., R. A. de Man, E. Fries, A. B. van Nunen, A. D. M. E. Osterhaus, and H. G. M. Niesters. 2002. The dynamics of mutations in the polymerase gene of hepatitis B virus during and after lamivudine treatment. J. Clin. Virol. 25:63-71. [DOI] [PubMed]
- 17.Pichoud, C., B. Seignères, Z. Wang, C. Trèpo, and F. Zoulim. 1998. Transient selection of a hepatitis B virus polymerase mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology 29:230-237. [DOI] [PubMed] [Google Scholar]
- 18.Richman, D. D. 2001. Antiretroviral activity of emtricitabine, a potent nucleoside reverse transcriptase inhibitor. Antiviral Ther. 6:83-88. [PubMed] [Google Scholar]
- 19.Rousseau, F., J. Kahn, M. Thompson, D. Mildvan, D. Shepp, J. Sommadossi, J. Delehanty, J. Simpson, L. Wang, J. Quinn, C. Wakeford, and C. van der Horst. 2001. Prototype trial design for rapid dose selection of antiretroviral drugs: an example with emtricitabine (Coviracil). J. Antimicrob. Chemother. 48:507-513. [DOI] [PubMed] [Google Scholar]
- 20.Si Ahmed, S. N., D. Tavan, C. Pichoud, F. Berby, L. Stuyver, M. Johnson, P. Merle, H. Abidi, C. Trépo, and F. Zoulim. 2000. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in patients treated by lamivudine for chronic hepatitis B. Hepatology 32:1078-1088. [DOI] [PubMed] [Google Scholar]
- 21.Stuyver, L., C. Van Geyt, S. De Gendt, G. Van Reybroeck, F. Zoulim, G. Leroux-Roels, and R. Rossau. 2000. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Virol. 38:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuyver, L., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Shinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 23.Van Geyt, C., S. De Gendt, A. Rombaut, A. Wyseur, G. Maertens, R. Rossau, and L. Stuyver. 1998. A line probe assay for hepatitis B virus genotypes, p. 139-145. In R. F. Schinazi, J. P. Sommadossi, and H. Thomas (ed.), Therapies of viral hepatitis. International Medical Press, London, United Kingdom.
- 24.Van Laethem, K., K. Van Vaerenbergh, J.-C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clerq, and A.-M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assay for detection of resistance in mixed human immunodeficiency virus-1 genotypic populations. J. Acquired Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 25.Yeh, C. T., R. N. Chien, C. M. Chu, and Y. F. Liaw. 2000. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology 31:1318-1326. [DOI] [PubMed] [Google Scholar]
- 26.Yuen, M. F., E. Sablon, C. H. Hui, H. J. Yuan, H. Decraemer, and C. L. Lai. 2001. Factors preceding hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology 34:785-791. [DOI] [PubMed] [Google Scholar]