Abstract
Multidrug-resistant opportunistic pathogens have become endemic to the veterinary hospital environment. Escherichia coli isolates resistant to 12 antibiotics were isolated from two dogs that were housed in the intensive care unit at The University of Georgia Veterinary Teaching Hospital within 48 h of each other. Review of 21 retrospective and prospective hospital-acquired E. coli infections revealed that the isolates had similar antibiotic resistance profiles, characterized by resistance to most cephalosporins, β-lactams, and the β-lactamase inhibitor clavulanic acid as well as resistance to tetracycline, spectinomycin, sulfonamides, chloramphenicol, and gentamicin. E. coli isolates with similar resistance profiles were also isolated from the environment in the intensive care unit and surgery wards. Multiple E. coli genetic types were endemic to the hospital environment, with the pulsed-field gel electrophoresis fingerprint identified among E. coli isolates from diseased animals and the hospital environment matching. The extended-spectrum cephalosporin resistance in these nosocomial E. coli isolates was attributed to the cephamycinase-encoding gene, blaCMY2. Chloramphenicol resistance was due in part to the dissemination of the florfenicol resistance gene, flo, among these isolates. Resistance encoded by both genes was self-transmissible. Although blaCMY2 and flo were common to the polyclonal, nosocomial E. coli isolates, there was considerable diversity in the genetic compositions of class 1 integrons, especially among isolates belonging to the same genetic type. Two or more integrons were generally present in these isolates. The gene cassettes present within each integron ranged in size from 0.6 to 2.4 kb, although a 1.7-kb gene cassette was the most prevalent. The 1.7-kb gene cassette contained spectinomycin resistance gene aadA5 and trimethoprim resistance gene dfrA17.
One of the great challenges of practicing veterinary medicine and surgery in tertiary-care facilities in the postantibiotic era is multidrug-resistant nosocomial infections. Control of Escherichia coli infections in veterinary medicine has become especially problematic due to the emergence of multiple-antibiotic-resistant E. coli in food animals and companion animals (3, 5, 43, 44, 61, 64, 67).
Although there is considerable information concerning the epidemiology and ecology of hospital-acquired infections in human medicine (17), little is known about nosocomial illnesses in veterinary hospitals or clinics (15, 18, 19, 30, 31, 58, 59). Most of what is known about hospital-acquired infections in veterinary medicine is primarily limited to a few retrospective studies (19, 30, 31, 59).
During the past 2 years, several dogs developed postoperative surgical wound or lower urinary tract infections during their stay in The University of Georgia Small Animal Veterinary Teaching Hospital (UGA-SVTH). E. coli was often isolated from the site of infection. This organism is an important pathogen of companion animals (4, 46). These isolates were unusual in that they all had the same antibiotic susceptibility pattern. A dog with extensive bite wounds died suddenly from suspected septic shock. He was housed in the intensive care unit (ICU) that nine days earlier had contained a dog with a hospital-acquired E. coli infection of a surgical incision. In addition to an Enterococcus sp., E. coli was isolated from the bite wounds of the dog that died, and this isolate also had the same antibiotic resistance pattern as the E. coli isolate from the dog previously housed in the same unit. Was a single E. coli clone responsible for these infections, or was the dissemination of a common resistance plasmid in the E. coli population making the isolates more likely to cause disease due to their resistance to the antibiotics commonly prescribed in the teaching hospital? In an attempt to address this question, we typed E. coli isolates by ERIC PCR, pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA (RAPD) analysis. In this report, we identify several E. coli clones associated with nosocomial E. coli infections at our institution.
MATERIALS AND METHODS
Bacterial strains.
The present study included 21 bacterial isolates obtained from urine and wounds of dogs that were patients at UGA-SVTH, 1 isolate from a dog that had never been a patient at UGA-SVTH, and 12 environmental samples from the ICU and surgery wards of UGA-SVTH. Bacteriological examination of the clinical samples was carried out as follows; swabs were plated on blood agar (Difco, Sparks, Md.) and MacConkey agar (Difco) at 37°C overnight. The environmental samples were obtained with three sterile sponges (3 × 1 cm) soaked in sterile saline solution; after sampling the sponges were placed in EC broth (Difco), incubated at 45°C overnight, and then plated onto MacConkey agar plates containing 32 μg of chloramphenicol per ml. Chloramphenicol was chosen since resistance to this drug is rather unusual, especially in light of its limited use in veterinary and human medicine. E. coli isolates were identified by standard procedures (9). All isolates were diluted in 15% glycerol and peptone water and stored at −70°C.
Antimicrobial susceptibility determination.
Antimicrobial MICs for E. coli isolates were determined with the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, Ohio) and were interpreted according to National Committee for Clinical Laboratory Standards (NCCLS) guidelines for broth microdilution methods (39-41). Susceptibility testing with the Sensititre system was performed according to the instructions of the manufacturer. The following antimicrobials were assayed: amikacin, amoxicillin-clavulanic acid, ampicillin, cefazolin, cefoxitin, ceftiofur, cephalothin, chloramphenicol, clindamycin, enrofloxacin, orbifloxacin, gentamicin, imipenem, sulfadimethoxine, tetracycline, ticarcillin, and trimethoprim-sulfamethoxazole. For each E. coli isolate, the disk diffusion method with florfenicol disks was performed by the method described by the NCCLS (39-41).
Characterization of class 1 integrons and antibiotic resistance genes associated with canine E. coli isolates.
Multidrug-resistant E. coli isolates were screened by PCR for the florfenicol resistance gene (flo) (27), the extended-spectrum cephalosporinase gene (blaCMY2) (67), and the class 1 integrase gene (intI1) (3). We characterized the gene cassette associated with the aatI integration site of class 1 integrons by PCR and sequenced the amplicon using primers specific for the conserved 5′ and 3′ sequences flanking the integration site (32). The transmissibility of extended-spectrum cephalosporin resistance was determined by bacterial conjugation with rifampin-resistant Salmonella enterica subsp. enterica serovar Typhimurium as a recipient in filter matings (48). This recipient strain is sensitive to all antibiotics except rifampin. Transconjugants were selected by plating the isolates used in the filter matings on brilliant green agar with rifampin and ampicillin. The antibiotic susceptibilities of the Salmonella transconjugants were determined as described in the preceding section.
Plasmid DNA was isolated from E. coli by alkaline lysis with a plasmid DNA extraction kit (QIAprep Spin Miniprep Kit; Qiagen, Valencia, Calif.). E. coli V517 served as a positive control for plasmid extraction. This E. coli isolate contains plasmids of high, intermediate, and low molecular weight (MW) (33). Plasmid DNA (10 μl) was separated on a 0.8% agarose gel containing 1× TAE (Tris-acetate-EDTA) at 100 V for 1 h and was visualized by staining with ethidium bromide (0.2 μg/ml). A supercoiled DNA ladder (Promega, Madison, Wis.) was used as an MW standard for determination of the MW of the plasmid(s). Plasmid or megabase DNA (PFGE) from the gels was blotted onto nylon membranes with a Bio-Rad (Hercules, Calif.) Vacuum Blotter (61). DNA-DNA hybridizations were done by the procedure of Sambrook et al. (49), with a temperature of 68°C used for hybridizations and washes. The blaCMY2-specific DNA probe was labeled with digoxigenin-labeled nucleotides by PCR (67), and membrane-bound probe was detected by the procedure of Bass et al. (3).
Molecular typing of clinical E. coli isolates.
E. coli isolates were typed by RAPD analysis with primer 1290 (38) and by ERIC PCR (60). Whole-cell template was prepared for RAPD analysis and ERIC PCR by the protocol of Hilton et al. (23). RAPD analysis and ERIC PCR were performed with an Idaho Technologies (Idaho Falls, Idaho) Rapidcycler hot-air thermocycler (65). The conditions for RAPD analyses were those described for the procedure of Maurer et al. (38). The program parameters for ERIC PCR with the hot-air thermocycler were as follows: (i) 94°C for 15 s; (ii) 94°C for 0 s, 52°C for 0 s, and 72°C for 15 s (slope = 2.0) for 30 cycles; and (iii) final extension at 72°C for 4 min. DNA was separated on a 1.5% agarose gel containing 1× TAE at 100 V for 1 h and was visualized by ethidium bromide staining (0.2 μg/ml) (49). A 100-bp ladder (GIBCO/BRL, Gaithersburg, Md.) was used as an MW standard for determination of the MWs of the PCR products.
For the typing of the bacteria by PFGE, agarose-embedded bacterial genomic DNA was digested with 10 U of restriction enzyme XbaI or BlnI overnight at 37°C, and DNA fragments were separated by PFGE (2) in a 1% PFGE agarose gel (Bio-Rad) with a CHEF DR-II electrophoretic apparatus (Bio-Rad). Electrophoresis was for 25 h with a voltage of 200 V and a linearly ramped pulse time of 2 to 40 s (2). Saccharomyces cerevisiae chromosomes (Boehringer Mannheim, Indianapolis, Ind.) served as MW markers for PFGE. The restriction enzyme XbaI has proved useful in the typing of E. coli (2) and other gram-negative bacteria (1, 45) by PFGE. For isolates with indistinguishable XbaI PFGE DNA patterns, other molecular typing methods were introduced to discern more subtle genetic differences among isolates (45): RAPD analysis (23, 25, 26), a second PFGE with BlnI as the restriction enzyme (57), or ERIC PCR (12).
Nucleotide sequence accession number.
The nucleotide sequences of PCR amplicons from canine E. coli isolates have been submitted to GenBank and given accession no. AF475279, AF475280, and AF475281.
RESULTS AND DISCUSSION
A 7-year-old male boxer was admitted to UGA-SVTH on 12 December 1999 for treatment of severe bite wounds to his neck, thorax, and abdomen inflicted during a dogfight. Five closed-suction surgical drains were implanted into the bite wounds after debridement and primary closure. Five days after admission to the hospital, the wounds were debrided a second time. Significant tissue necrosis had occurred. The dog died suddenly postoperatively from suspected septic shock. In addition to an Enterococcus sp., E. coli (clinical isolate 25055) was isolated from samples taken from the bite wounds during the second surgery (Tables 1 and 2). The organism was resistant to most cephalosporins, β-lactams and β-lactamase inhibitors, chloramphenicol, spectinomycin, tetracycline, gentamicin, and enrofloxacin, as determined by broth microdilution methods. The only drugs to which the isolate was susceptible were amikacin and imipenem. A similar multidrug-resistant E. coli isolate had been isolated from a dog that had developed a postoperative surgical wound infection 9 days earlier. Both animals had been housed in the ICU at UGA-SVTH within 48 h of each other.
TABLE 1.
Bacterial strains
| Isolatea | Date (mo/day/yr) | Sourceb | Sexc | Age (yr) | Sited | Genetic typee
|
|||
|---|---|---|---|---|---|---|---|---|---|
| PFGE
|
ERIC PCR | RAPD analysis | |||||||
| BlnI | XbaI | ||||||||
| 39737 | 4/10/00 | Springer spaniel | F | 12 | Wounds | A1 | H1.1 | II | Q |
| 22-2 | 8/22/00 | Storage cart 2 | NA | NA | NA | A1 | H1.1 | JJ | R |
| 29 | 8/22/00 | Cage no. 3 | NA | NA | NA | A1 | H1.1 | II | R |
| 34 | 8/22/00 | Drain 3 runs | NA | NA | NA | A1 | H1.1 | II | R |
| 53 | 8/22/00 | ICU floor | NA | NA | NA | A1 | H1.1 | KK | R |
| 56 | 8/22/00 | ICU floor | NA | NA | NA | A1 | H1.1 | KK | R |
| 22949 | 1/3/00 | Labrador | M | 14 | Wounds | A2.2 | H1.2 | II | S |
| 4479 | 8/3/00 | Labrador | F | 15 | Bile | A2.2 | H1.2 | LL | T |
| 38 | 8/22/00 | Small hydrobath | NA | NA | NA | A2.1 | H1.3 | MM | U |
| 4517A | 8/6/98 | Great Pyrenees | M | 3 | Wounds | A2.3 | I | NN | V |
| 4517B | 8/6/98 | Great Pyrenees | M | 3 | Wounds | A2.3 | I | NN | V |
| 22255 | 12/17/98 | Mixed | M | ? | Wounds | A2.3 | I | OO | V |
| A1-72 | 7/5/00 | Cocker spaniel | F | 9 | CSF | B1.2 | J | PP | S |
| 1745 | 7/19/00 | Great Dane | M | 10 | Urine | B1.2 | J | W | |
| 32 | 8/22/00 | Drain 1 runs | NA | NA | NA | B1.2 | J | RR | R |
| 27315 | 1/31/00 | Corgi | M | 8 | Urine | B1.1 | K | KK | X |
| 40362 | 4/12/99 | Mixed | M | 6 | Urinec | B1.1 | L | NN | Y |
| 42614B | 4/21/00 | Boxer | F | 4 | Urine | D | M | SS | Z |
| 1888 | 7/17/00 | Samoyed | M | 2 | Wounds | G | N | JJ | AA |
| 18813 | 10/31/00 | Spitz | F | 11 | A. sac | G | N | JJ | AA |
| 22559 | 12/22/99 | Boxer | M | 7 | Wounds | E | H1.1 | II | U |
| 25055a | 1/18/00 | Labrador | M | 14 | Wounds | E | H1.1 | II | U |
| 25055b | 1/18/00 | Labrador | M | 14 | Wounds | E | H1.1 | II | U |
| 29610f | 2/14/00 | Mixed | F | 11 | Urine | C | O | OO | BB |
| 12279 | 9/30/99 | Mixed | F | 12 | Urine | C | O | TT | CC |
| 21 | 8/22/00 | Storage cart 1 | NA | NA | NA | F | P | JJ | R |
| 42614A | 4/21/00 | Boxer | F | 4 | Urine | B1.1 | H1.2 | II | Q |
| 18397f,g | 11/16/98 | Saluki | F | 9 | RT | —h | — | UU | DD |
| 23120f,g | 12/12/00 | Mixed | F | 1 | Urine | — | — | VV | EE |
| 21411f,g | 11/16/00 | Pomeranian | F | 8 | Urine | — | — | VV | FF |
| 37 | 8/22/00 | Big hydrobath | NA | NA | NA | — | — | VV | GG |
| 55-1 | 8/22/00 | ICU floor | NA | NA | NA | — | — | WW | HH |
| 55-2 | 8/22/00 | ICU floor | NA | NA | NA | — | — | WW | HH |
| 55-3 | 8/22/00 | ICU floor | NA | NA | NA | — | — | LL | HH |
E. coli organisms were isolated from dogs that developed infections >48 h after admission to UGA-SVTH.
All animals but the one from which isolate 29610 was obtained had been patients at UGA-SVTH.
F, female; M, male; NA, not available.
Site of infection was usually associated with an indwelling medical device. s, surgical drain; c, urinary catheter; CSF, cerebrospinal fluid; RT, reproductive tract; A. sac, anal sac.
We chose PFGE, RAPD analysis, and ERIC PCR for typing of the E. coli isolates. For PFGE, the restriction enzymes XbaI and BlnI were selected for use for the typing of the bacterial isolates. RAPD analysis involved the use of a 10-mer oligonucleotide, primer 1290, as the typing primer. An alphabetical designation was assigned to each distinct DNA fingerprint. Letters highlighted in boldface identify isolates with the same pattern by PFGE with either the XbaI or BlnI restriction enzyme. Since no interpretative criteria have been developed for analysis of either RAPD or ERIC PCR patterns, DNA patterns with one or more band differences were given a different alphabetical designation.
E. coli organisms isolated from samples submitted from regional veterinary practitioners to The Athens Diagnostic Laboratory for culture.
Isolates obtained through submission by a clinician outside UGA-SVTH.
—, isolates were recalcitrant to typing by PFGE.
TABLE 2.
Antibiotic resistance profiles of nosocomial E. coli isolates
| Isolate | Genetic typea | Antibiotic resistance profileb | intI | Antibiotic resistance genesc
|
||||
|---|---|---|---|---|---|---|---|---|
| Class 1 integron
|
Cephamycinase
|
flo | ||||||
| Cassette(s) size (kb) | Genes | blaCMY2 | Molecular size (kb) of fragment generated by PFGEd | |||||
| 39737 | 1A | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri | + | 1.7, 0.80 | aadA5, dfrA17 | + | − | |
| 22-2 | 1A | ESCp, Chl, Flq, Spc, Tet I, Sul, Tri, Flor | + | 1.7, 0.90 | aadA5, dfrA17 | + | − | |
| 29 | 1A | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri | + | 1.7, 0.60 | aadA5, dfrA17 | + | 25 | − |
| 34 | 1A | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.7, 0.90 | aadA5, dfrA17 | + | − | |
| 53 | 1A | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri | + | 1.5, 0.75 | + | − | ||
| 56 | 1A | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor I | + | 1.5, 0.75 | + | − | ||
| 22949 | 1B | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 1.4, 0.70 | + | + | ||
| 4479 | 1B | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 1.3, 0.75 | + | + | ||
| 38 | 1B | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 1.2, 0.60 | + | 185 | + | |
| 4517A | 2 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.7, 0.75 | aadA5, dfrA17 | + | 250 | + |
| 4517B | 2 | ESCp, Chl, Flq, Spc, Tet, Sul, Tri, Flor | + | 1.7, 0.75 | aadA5, dfrA17 | + | + | |
| 22255 | 2 | ESCp, Chl, Flq, Spc, Tet, Sul, Tri, Flor | + | 0 | + | + | ||
| A1-72 | 3 | ESCp, Chl I, Flq, Spc I, Sul, Gen, Flor, Tet | − | 0 | + | 100 | + | |
| 1745 | 3 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 0.90 | + | − | ||
| 32 | 3 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.7, 0.90 | aadA5, dfr17 | + | − | |
| 27315 | 4 | ESCp, Chl, Flq, Spc I, Tet, Flor | + | 0 | + | 140 | + | |
| 40362 | 5 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | − | 0 | + | + | ||
| 42614B | 6 | Sul | − | 0 | − | 280, 115 | − | |
| 1888 | 7 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.7, 1.1, 0.75 | aadA5, dfrA17 | + | 30, 27 | + |
| 18813 | 7 | ESCp, Chl, Flor, Amk I | + | 0 | + | + | ||
| 22559 | 8 | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 1.4, 0.70 | + | + | ||
| 25055a | 8 | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 0 | + | + | ||
| 25055b | 8 | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 1.3, 0.70 | + | + | ||
| 29610 | 9 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.3, 0.70 | + | + | ||
| 12279 | 9 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.2, 0.60 | − | + | ||
| 21 | 10 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor I | + | 1.7, 0.90 | aadA5, dfr17 | + | 29 | − |
| 42614A | 11 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | − | 0 | + | + | ||
| 18397 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 2.4, 1.2 | + | + | |||
| 23120 | ESCp, Chl, Flq, Spc I, Tet, Gen, Sul, Tri, Flor | + | 0 | + | + | |||
| 21411 | ESCp, Chl, Flq, Spc, Tet, Gen, Sul, Tri, Flor | + | 1.4, 0.70 | + | + | |||
| 37 | ESCp, Chl, Flq, Spc, Tet, Sul, Tri | + | 0 | + | − | |||
| 55-1 | ESCp, Chl, Flq, Tet, Sul, Tri, Flor I | + | 0 | + | − | |||
| 55-2 | ESCp, Chl, Flq, Tet, Sul, Tri, Flor I | + | 0 | + | − | |||
| 55-3 | ESCp, Chl, Flq, Spc I, Tet, Sul, Tri, Flor | + | 0 | + | − | |||
A numerical designation was assigned to each distinct DNA fingerprint generated by PFGE with both restriction enzymes XbaI and BlnI. Isolates with same number have identical PFGE patterns with either restriction enzyme.
ESCp, extended-spectrum cephalosporin resistance, including resistance to the cephalosporins (cefazolin, cefoxitin, cephalothin and ceftioufur), β-lactams (amoxicillin, ampicillin and ticarcillin), and the β-lactam inhibitor clavulanic acid; Flq, fluoroquinolones (enrofloxacin and orbifloxacin). The bacterial isolates were also resistant to chloramphenicol (Chl), gentamicin (Gen) spectinomycin (Spc), sulfonamides (Sul), tetracycline (Tet), and trimethoprim (Tri).
Bacterial isolates were screened for the presence of class 1 integrons by PCR by using the integrase gene intI as a marker for this genetic element. A second PCR was done with primers specific for conserved sequences flanking the integron integration site aatI. This PCR amplifies a gene cassette(s) present within aatI (32). PCR amplicons were sequenced, and the nucleotide or translated amino acid sequences were queried against the GenBank DNA database at the National Center for Biotechnology Information for matches. The genes and the corresponding integron cassette size(s) are highlighted in boldface. Other antibiotic resistance genes for which the isolates were surveyed in this study included blaCMY2 (67) and flo (27).
Genetic mapping of blaCMY2 was done for canine E. coli isolates representative of the different genetic types by DNA-DNA hybridization of PFGE generated with restriction endonuclease XbaI and hybridized with PCR-generated DNA probe specific for blaCMY2. The molecular sezis of XbaI DNA fragment(s) that hybridized with blaCMY2 probe are given.
A retrospective and prospective study of clinical submissions to The Athens Diagnostic Laboratory from UGA-SVTH identified 18 E. coli isolates from dogs with the same patterns of resistance to 12 antibiotics reported earlier by broth microdilution methods. In several of these cases, E. coli infection was associated with placement of a surgical wound drain or an indwelling urinary catheter. A thorough environmental sampling of the surgery rooms, surgery wards, and ICU for microorganisms was done. E. coli organisms were isolated from 42 of 55 locations surveyed. Twelve of these E. coli organisms exhibited the multidrug resistance profiles observed for clinical isolates obtained from materials submitted from the veterinary hospital. Are the multidrug-resistant E. coli infections observed in dogs admitted to UGA-SVTH attributed to a clone that is endemic to the hospital?
Genetic diversity in E. coli isolates associated with nosocomial infections in dogs.
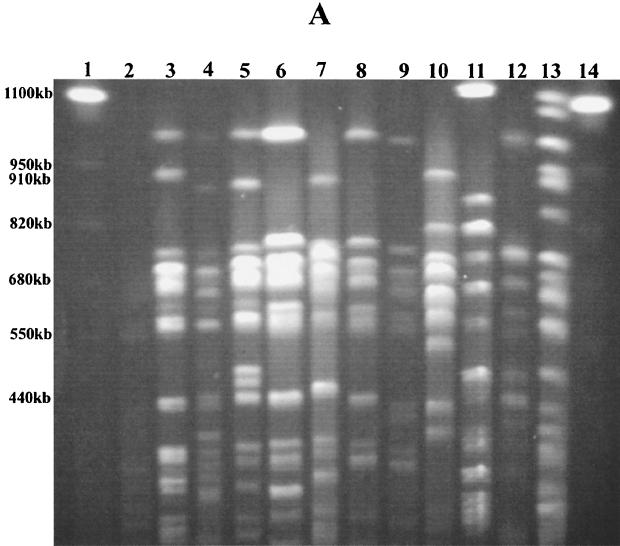
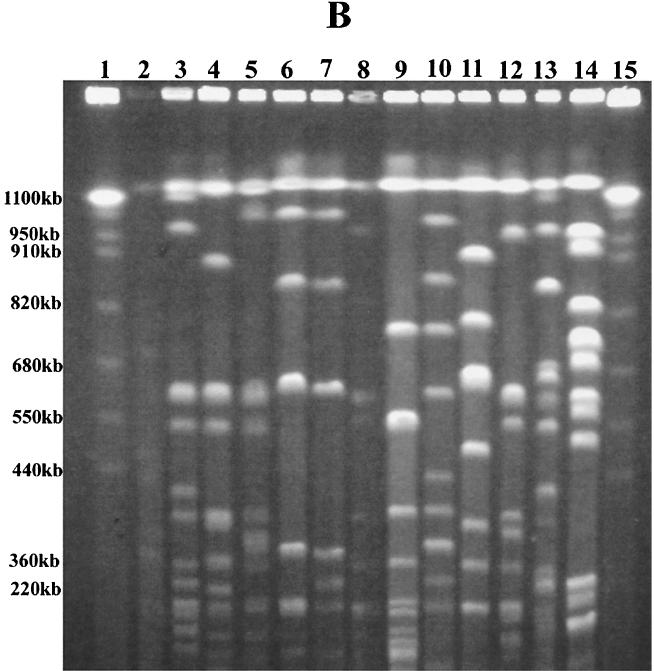
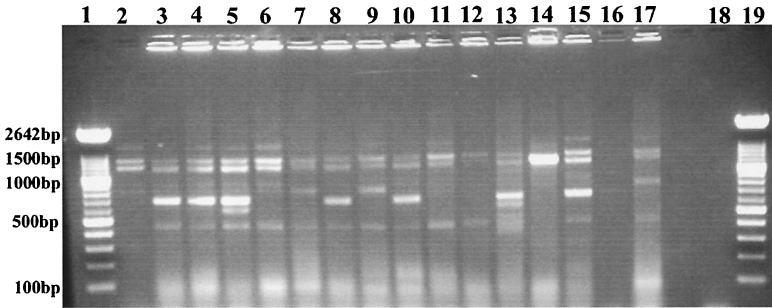
Nine distinct DNA patterns were observed among the 34 E. coli isolates typed by PFGE with XbaI (Table 1; Fig. 1A). Seven E. coli isolates could not be typed by PFGE due to problems with degradation of the genomic DNA during preparation of the agarose plugs. Other laboratories have noted similar problems with typing by PFGE (22, 35, 36). Isolates with seven or more different DNA fragments were assigned to a specific XbaI PFGE genetic type (types H to P), as recommended by Tenover et al. (56). Forty percent (7 of 18) of the clinical E. coli isolates were XbaI PFGE genetic type H. Half (6 of 12) of the environmental isolates were also XbaI PFGE DNA type H. Additional differences of two to three DNA fragments among the isolates of PFGE type H allowed further differentiation of these related E. coli isolates into subgroups H1.1 to H1.3. Using PFGE with a second restriction enzyme, BlnI, we were unable to distinguish among E. coli isolates that had earlier produced similar or identical patterns by PFGE with XbaI (Table 1; Fig. 1B). Seven distinct PFGE patterns (DNA patterns A to G) were observed with restriction enzyme BlnI. With this second restriction enzyme, isolates of XbaI PFGE type H could be further discriminated into three genetic types. By ERIC PCR and RAPD analysis with primer 1290, we could further discriminate several of these isolates, although in most cases we were able to identify only minor band differences in the patterns generated by either of the methods (Fig. 2 and 3). We identified the same E. coli genetic type among organisms isolated either from different animals admitted to the veterinary hospital (isolates 4517 and 22255, isolates 1888 and 18813, and isolates 22559 and 25055) or from a hospitalized animal and the veterinary clinic environment (isolates 29737, 29, and 34 and isolates 1745 and 32, respectively). Instead of identifying one multidrug-resistant clone that was endemic to the hospital environment, as has been reported for vancomycin-resistant enterococci (29), we identified several multidrug-resistant clones in the hospital environment. Others have made similar observations for gram-negative organisms that cause nosocomial infections in humans (21, 51, 53). Are we dealing with dissemination of a common plasmid in the hospital environment responsible for resistance to 11 antibiotics, including most of the broad-spectrum cephalosporins?
FIG. 1.
E. coli genetic types identified by PFGE. (A) Patterns obtained by PFGE with XbaI. Lanes 1 and 14, S. cerevisiae DNA standards (BioWhittaker Molecular Applications, Rockland, Maine); lane 3, PFGE pattern H1 (isolate 29); lane 4, H1.2 (isolate 42614A); lane 5, H1.4 (isolate 38); lane 6, I (isolate 4517A); lane 7, J (isolate A1-72); lane 8, K (isolate 27315); lane 9, L (isolate 40362); lane 10, M (isolate 42614B); lane 11, N (isolate 1888); lane 12, O (isolate 29610); and lane 13, P (isolate 21). (B) Patterns obtained by PFGE with BlnI. Lanes 1 and 15, S. cerevisiae DNA standards (BioWhittaker Molecular Applications); lane 3, PFGE pattern A1 (isolate 29); lane 4, A2.1 (isolate 38); lane 5, A2.2 (isolate 4479); lane 6, A2.3 (isolate 4517A); lane 7, B1 (isolate 27315); lane 8, B1.1 (isolate 42614A); lane 9, B1.2 (isolate A1-72); lane 10, C (isolate 29610); lane 11, D (isolate 42614B); lane 12, E (isolate 22559); lane 13, F (isolate 1888); and lane 14, G (isolate 21). E. coli K-12 LE392 in panels A and B (lanes 2) served as an internal control for the reproducibility of every PFGE run. Alphabetical designations were assigned to PFGE patterns with seven or more band differences (56). The numbers represent four to six band differences within a PFGE pattern, and the numbers following decimal points designate slight genetic differences of one to three bands.
FIG. 2.
E. coli genetic types identified by ERIC PCR. Lanes 1 and 20, 100-bp ladder (Roche Molecular Biochemicals, Indianapolis, Ind.); lane 3, ERIC pattern II (isolate 29); lane 4, RR (isolate 32); lane 5, LL (isolate 4479); lane 6, NN (isolate 4517A); lane 7, VV (isolate 37); lane 8, KK (isolate 56); lane 9, SS (isolate 42614B); lane 10, QQ (isolate 1745); lane 11, TT (isolate 12279); lane 12, OO (isolate 29610); lane 13, JJ (isolate 21); lane 14, UU (isolate 18397); lane 15, PP (isolate A1-72); lane 16, MM (isolate 38); lane 17, WW (isolate 55-1); lane 18, empty; and lane 19, no-DNA control. E. coli K-12 LE392 (lane 2) served as an internal control for the reproducibility of every ERIC PCR run. Since no interpretative criteria have been developed for analysis of the patterns obtained by ERIC PCR, DNA patterns with one or more band differences were given a different alphabetical designation.
FIG. 3.
E. coli genetic types identified by RAPD analysis with primer 1290. Lanes 1 and 22, 100-bp ladder (Roche Molecular Biochemicals); lane 2, RAPD 1290 pattern R (isolate 29); lane 3, FF (isolate 21411); lane 4, T (isolate 4479); lane 5, EE (isolate 23120); lane 6, U (isolate 22559); lane 7, Z (isolate 42614B); lane 8, Y (isolate 40362); lane 9, S (isolate A1-72); lane 10, GG (isolate 37); lane 11, HH (isolate 55-1); lane 12, V (isolate 4517A); lane 13, AA (isolate 1888); lane 14, Q (isolate 39737); lane 15, DD (isolate 18397); lane 16, X (isolate 27315); lane 17, BB (isolate 29610); lane 18, CC (isolate 12279); lane 19, W (isolate 1745); and lane 21, no-DNA control. E. coli K-12 LE392 (lane 20) served as an internal control for reproducibility of every RAPD analysis run. Since no interpretative criteria have been developed for analysis of RAPD PCR patterns, DNA patterns with one or more band differences were given a different alphabetical designation.
Antimicrobial susceptibility and drug resistance genes in nosocomial E. coli isolates associated with infections in dogs.
Nosocomial E. coli isolates were resistant to most cephalosporins including ceftiofur, cephalothin, and ceftriaxone, as well as to the β-lactams ampicillin and amoxicillin and the β-lactamase inhibitor clavulanic acid. This broad spectrum of resistance to β-lactams, cephalosporins, and β-lactamase inhibitors is a feature common to the ampC class of cephamycinases (11). One particular ampC-like gene, blaCMY2, was recently detected in ceftriaxone-resistant E. coli isolates of animal origin (67). We examined our canine isolates for the presence of this gene.
Thirty-two of 34 isolates screened by PCR were positive for the cephamycinase gene (blaCMY2). There was 99.9% identity between the nucleotide sequence of one of the PCR amplicons (GenBank accession no. AF475279) from a canine isolate and the published sequence of the blaCMY2 gene (67). The blaCMY2 gene is closely related to the Citrobacter freundii chromosomal ampC gene (66) and to the plasmid-associated ampC gene present in S. enterica subsp. enterica serovar Typhimurium isolates (16) and human and animal E. coli isolates (64, 67).
Narrow-spectrum parenteral cephalosporins are used extensively for prophylaxis in cats and dogs with surgical wounds (8). At UGA-SVTH, cefazolin is the cephalosporin most commonly used parenterally. Oral cephalosporins, such as cephalexin, are used for the treatment of skin and urinary tract infections caused by susceptible organisms. Other applications include the treatment of abscesses and wound infections caused by susceptible organisms in dogs and cats. Expanded-spectrum cephalosporins are indicated for mixed infections with anaerobes (aspiration pneumonia, severe bite wound infections, gangrene, peritonitis, pleuritis) and prophylaxis in colonic or perineal surgery. Broad-spectrum cephalosporins are used only for treatment of infections caused by otherwise resistant bacteria in cats and dogs. It has been estimated that 13% of the E. coli isolates involved in scours in calves are resistant to broad-spectrum cephalosporins, most likely due to hyperproduction of the chromosomally encoded AmpC β-lactamase (10).
Another unusual resistance phenotype reported among these canine and environmental E. coli isolates was resistance to the phenicols chloramphenicol and the veterinary analog florfenicol. This resistance was attributed to widespread dissemination of the cmlA homologue flo among gram-negative bacteria (7, 13, 14, 24, 27, 28, 61). Twenty of 34 E. coli isolates possessed the flo florfenicol resistance gene.
The Food and Drug Administration approved the veterinary use of the fluorinated analog of chloramphenicol, florfenicol, for the treatment of bovine respiratory disease in 1996. Florfenicol does not cause irreversible aplastic anemia, and it is not susceptible to inactivation by chloramphenicol transacetylases. Its mechanism of action is very similar to that of chloramphenicol. Although the Food and Drug Administration has approved the use of florfenicol only for the treatment of respiratory disease in cattle caused by highly susceptible bacteria such as Pasteurella and Haemophilus, other possible future uses of florfenicol have been documented, such as for the treatment of mastitis in cattle (63), to obtain reductions in rates of morbidity and mortality from infections caused by Actinobacillus pleuropneumoniae in pigs (J. A. Jackson et al., Proc. 15th Int. Pig VET Congr., abstr. P187, 1998), and for the treatment of furunculosis in fish (50). Of the phenicols, only chloramphenicol is used at UGA-SVTH, but it is used infrequently.
We were able to demonstrate the transfer of extended-spectrum cephalosporin and florfenicol resistance and the antibiotic resistance genes blaCMY2 and flo, respectively, from canine E. coli isolate 4517A to rifampin-resistant S. enterica subsp. enterica serovar Typhimurium at a frequency of 10−6. Other antibiotics to which resistance was transferred to the recipient included gentamicin, spectinomycin, and sulfadimethoxime. Navarro et al. (42) also reported the transfer of resistance to the same antibiotics to recipient E. coli isolates from Salmonella, Klebsiella pneumoniae, Proteus mirabilis, and E. coli isolates that possessed the blaCMY2 cephamycinase gene (42). The identity of the chloramphenicol resistance gene was not determined in that study. Conjugative R plasmids have been reported for canine E. coli isolates that confer resistance to four or more antibiotics including ampicillin, tetracycline, chloramphenicol, sulfonamides, and streptomycin (43). Although a specific antibiotic resistance gene was never identified, other investigators (37) reported on E. coli isolates from dogs treated with β-lactams which possessed transferable cephamycinases with the spectra of activity, MWs, and pIs characteristic of those of BlaCMY2. There have been other reports on the transmissibility of antibiotic resistance genes (blaCMY2 or flo) in Salmonella (42, 64, 67), E. coli (13, 42, 64, 67), K. pneumoniae (12, 42), P. mirabilis (42), and Vibrio cholerae (24). The cephamycinase gene described by Winokur et al. (64) appears to reside on a common plasmid shared between Salmonella and E. coli isolates from cattle. Since both blaCMY2 and flo are transferable, is the multidrug resistance present in polyclonal, nosocomial E. coli isolates due to the dissemination of a common, conjugative R plasmid in the veterinary teaching hospital?
Southern analysis was used to determine if the blaCMY2 cephamycinase gene mapped to a common XbaI DNA fragment, indicating dissemination of a common plasmid among these nosocomial isolates. We found that blaCMY2 mapped to a diverse array of XbaI DNA fragments in these isolates (Table 2). Canine E. coli isolates contained a diverse array of high-, intermediate-, and low-molecular-mass plasmids that ranged in size from 2 to 20 kb (Fig. 4). A 6.6-kb plasmid was common to 8 of 22 isolates examined (isolates 22, 34, 53, 18397, 22559, 25055a, 25055b, and 39737). When hybridized with a probe specific for blaCMY2 DNA, only the 20-kb plasmid in E. coli 4479 contained the blaCMY2 cephamycinase gene (data not shown), suggesting that this resistance gene maps to the chromosome or plasmids with higher MWs that cannot be isolated by the procedure used in this study. A similar observation was made with regard to the mapping of the flo florfenicol resistance gene in bovine E. coli isolates (61), suggesting that these resistance genes are being disseminated not by a common plasmid but, rather, by a transposable element (66). However, extended-spectrum cephalosporin resistance could have emerged in the hospital through the acquisition of a common, conjugative plasmid, but over time this plasmid may have changed as the plasmid acquired a transposon(s) or an integron(s) in this environment. Sequencing of the regions upstream and downstream of blaCMY2 will determine whether we are dealing with a transposable element that left the original plasmid from which the extended-spectrum cephalosporin resistance originated or whether this plasmid has undergone rapid genetic changes since its introduction into the hospital environment.
FIG. 4.
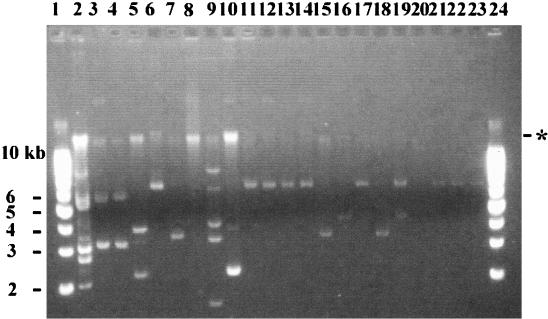
Plasmid profiles of canine E. coli isolates. Lanes 1 and 24, supercoiled DNA ladder; lane 2, V517, plasmid control strain; lane 3, isolate A1-72; lane 4, isolate 1745; lane 5, isolate 1888; lane 6, isolate 4479; lane 7, isolate 4517A; lane 8, isolate 12279; lane 9, isolate 18397; lane 10, isolate 18813; lane 11, 22559; lane 12, isolate 25055a; lane 13, isolate 25055b; lane 14, isolate 25055a; lane 15, isolate 27315; lane 16, isolate 29610; lane 17, isolate 39737; lane 18, isolate 40362, lane 19, isolate 42614A; lane 20, isolate 42614B; lane 21, isolate 22; lane 22, isolate 34; and lane 23, isolate 53. The asterisk indicates the position of chromosomal DNA on the gel.
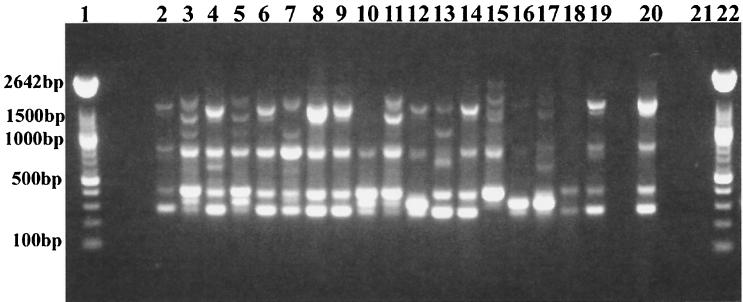
In addition to extended-spectrum cephalosporin and chloramphenicol resistance, the nosocomial E. coli isolates were also resistant to the antibiotics spectinomycin, sulfonamide, and gentamicin. Resistance to multiple antibiotics, especially to spectinomycin and sulfonamides, is often associated with integrons, genetic elements that acquire and trade antibiotic resistance genes (55). Recombination involves the integrase IntI1, integration site attI, and a 59-bp element common to genes for antibiotic resistance present within the integron (54, 55). A feature of class 1 integrons, in addition to intI1, is the sul1 sulfonamide resistance gene (55). Thirty of 34 nosocomial isolates in the present study possessed the intI1 integrase gene. Among the members of the family Enterobacteriaceae, the incidence of class 1 integrons was highest in E. coli and Salmonella isolates and was associated with multidrug resistance (20). To further characterize the integrons of the nosocomial E. coli isolates, PCR was used to amplify the gene cassette(s) associated with class 1 integrons (32). Four intI1-positive E. coli isolates produced no PCR amplicon, suggesting that the attI site of the integron did not contain a gene cassette, while two or more integrons were generally present in the remaining isolates that were positive for class 1 integrase (Fig. 5; Table 2). “Empty” class 1 integrons have been reported for other gram-negative bacteria (34, 52). The gene cassettes present within each integron ranged in size from 0.6 to 2.4 kb, although a 1.7-kb gene cassette was the most prevalent (n = 8). This 1.7-kb gene cassette was found in E. coli isolates of five different genetic types, suggesting that this integron is disseminated via a common genetic element, possibly a conjugative plasmid. Sequencing of the 1.7-kb PCR amplicon revealed the identities of two antibiotic resistance genes, dfrA17 and aadA5, genes that confer resistance to trimethoprim and spectinomycin, respectively (GenBank accession no. AF475280 and AF475281). The same integron was reported in a human E. coli isolate associated with a urinary tract infection (62).
FIG. 5.
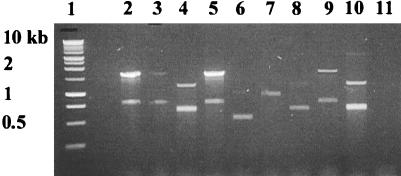
Gene cassette(s) in class 1 integrons of nosocomial E. coli. The gene cassette(s) present within the class 1 integron integration site (attI) was amplified by PCR with oligonucleotide primers specific for conserved sequences 5′ and 3′ of the attI site (32). PCR amplifications were done with canine E. coli isolates positive for the intI1class 1 integrase gene, a marker for class 1 integrons. Lanes 2 to 10, canine E. coli isolates with representative PCR amplicons associated with class 1 integrons in nosocomial isolates; lane 1, 1-kb ladder (Roche Molecular Biochemicals); lane 2, E. coli isolate 1745; lane 3, isolate 1888; lane 4, isolate 4479; lane 5, isolate 4517A; lane 6, isolate 12279; lane 7, isolate 18397; lane 8, isolate 21411; lane 9, isolate 39737; lane 10, isolate 38; and lane 11, no-DNA control.
To eradicate this problem from the veterinary teaching hospital, it was important to implement an effective infection control program (17, 47), to limit contact between affected patients (58), and to institute strict guidelines concerning the judicious use of antibiotics in the hospital. As is evident from our survey of the hospital environment, it is also important to thoroughly clean and disinfect affected areas (6, 58). A regular cleaning schedule was implemented, and we also have in place a program of monitoring for nosocomial infections (17). These measures have reduced the incidence of resistant E. coli isolates in the veterinary hospital.
Acknowledgments
We thank Paula Barlett and Kate Pennick for excellent technical support and undergraduate students Alex Gratz and Rosana Odeh for help with the environmental monitoring.
This work was possible thanks to a grant from UGA-SVTH and the full cooperation of clinicians and technical staff. J.J.M. was supported by USDA NRICGP grant 99-35212-8680.
REFERENCES
- 1.Baquar, N., A. Burnens, and J. Stanley. 1994. Comparative evaluation of molecular typing of strains from a national epidemic due to Salmonella brandenburg by rRNA gene and IS200 probes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:1876-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, L., C. A. Liebert, M. D. Lee, D. G. White, A. O. Summers, S. G. Thayer, and J. J. Maurer. 1999. The incidence and characterization of integrons, genetic elements associated with multiple drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L. 1999. Escherichia coli as a pathogen in dogs and cats. Vet. Res. 30:285-298. [PubMed] [Google Scholar]
- 5.Bischoff, K. M., D. G. White, P. F. McDermott, S. Zhao, S. Gaines, J. J. Maurer, and D. J. Nisbet. 2001. Characterization of chloramphenicol resistance in β-hemolytic Escherichia coli associated with diarrhea in neonatal swine. J. Clin. Microbiol. 40:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerlin, P., S. Eugster, F. Gaschen, R. Straub, and P. Schwalder. 2001. Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet. Microbiol. 82:347-359. [DOI] [PubMed] [Google Scholar]
- 7.Bolton, L. F., L. Kelly, M. D. Lee, P. F. Cray, and J. J. Maurer. 1999. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boothe, D. M. 2001. Treatment of bacterial infections, p. 217-219. In D. M. Boothe (ed.), Small animal clinical pharmacology and Therapeutics. The W. B. Saunders Company, Aharcourt Health Sciences Company, Philadelphia, Pa.
- 9.Bopp, C. A., F. W. Brenner, J. G. Wells, and N. A. Stockbine. 1999. Escherichia, Shigella, and Salmonella, p. 459-474. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 10.Bradford, P. A., P. J. Petersen, I. M. Fingerman, and D. G. White. 1999. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J. Antimicrob. Chemother. 44:607-610. [DOI] [PubMed] [Google Scholar]
- 11.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, C. G., A. N. Kravetz, C. Dendy, G. Wang, K. D. Tyler, and W. M. Johnson. 1998. Investigation of the 1994-5 Ukrainian Vibrio cholerae epidemic using molecular methods. Epidemiol. Infect. 121:15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloeckaert, A., S. Baucheron, and E. Chaslus-Dancla. 2001. Nonenzymatic chloramphenicol resistance mediated by IncC plasmid R55 is encoded by a floR gene variant. Antimicrob. Agents Chemother. 45:2381-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloeckaert, A., K. Sidi Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemther. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colahan, P. T., L. C. Peyton, M. R. Connelly, and R. Peterson. 1984. Serratia spp. infection in 21 horses. J. Am. Vet. Med. Assoc. 185:209-211. [PubMed] [Google Scholar]
- 16.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, E. Ribot, P. C. Iwen, P. A. Bradford, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with ampC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 17.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, J. G., C. M. Beaucage, C. A. Folta, and G. W. Thornton. 1981. Nosocomial transmission of Serratia marcescens in a veterinary hospital due to contamination by benzalkonium chloride. J. Clin. Microbiol. 14:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glickman, L. T. 1981. Veterinary nosocomial (hospital-acquired) Klebsiella infections. J. Am. Vet. Med. Assoc. 179:1389-1392. [PubMed] [Google Scholar]
- 20.Goldstein, C., M. D. Lee, S. Sanchez, C. R. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and normal flora bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyot, A., S. P. Barrett, E. J. Threlfall, M. D. Hampton, and T. Cheasty. 1999. Molecular epidemiology of multi-resistant Escherichia coli. J. Hosp. Infect. 43:39-48. [DOI] [PubMed] [Google Scholar]
- 22.Hielm, S., J. Bjorkroth, E. Hyytia, and H. Korkeala. 1998. Genomic analysis of Clostridium botulinum group II by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 64:703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilton, A. C., J. G. Banks, and C. W. Penn. 1997. Optimization of RAPD for fingerprinting Salmonella. Lett. Appl. Microbiol. 24:243-248. [DOI] [PubMed] [Google Scholar]
- 24.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson, C. R., M. Garcia, R. K. Gast, and J. J. Maurer. 2001. Determination of close genetic relatedness of the major Salmonella enteritidis phage types by pulsed-field gel electrophoresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis. 45:875-886. [PubMed] [Google Scholar]
- 26.Hudson, C. R., C. Quist, M. D. Lee, K. Keyes, S. V. Dodson, C. Morales, S. Sanchez, D. G. White, and J. J. Maurer. 2000. The genetic relatedness of Salmonella from nondomestic birds in the southeastern United States. J. Clin. Microbiol. 38:1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keyes, K., C. Hudson, J. J. Maurer, S. G. Thayer, D. G. White, and M. D. Lee. 2000. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 44:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, E. H., and T. Aoki. 1996. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen. Pasteurella piscisida. Microbiol. Immunol. 40:665-669. [DOI] [PubMed] [Google Scholar]
- 29.Kim, W., R. A. Weinstein, and M. K. Hayden. 1999. The changing molecular epidemiology and establishment of endemicity of vancomycin resistance in enterococci at one hospital over a 6-year period. J. Infect. Dis. 179:163-171. [DOI] [PubMed] [Google Scholar]
- 30.Koterba, A., J. Torchia, C. Silverthorne, R. Ramphal, A. M. Merritt, and J. Manucy. 1986. Nosocomial infections and bacterial antibiotic resistance in a university equine hospital. J. Am. Vet. Med. Assoc. 189:185-191. [PubMed] [Google Scholar]
- 31.Kruth, S. A., J. F. Prescott, M. K. Welch, and M. H. Brodsky. 1989. Nosocomial diarrhea associated with enterotoxigenic Clostridium perfringens infection in dogs. J. Am. Vet. Med. Assoc. 195:331-334. [PubMed] [Google Scholar]
- 32.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1994. PCR mapping integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macrina, F. L., D. J. Kopecko, K. R. Jones, D. J. Ayers, and S. M. McCowen. 1978. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1:417-420. [DOI] [PubMed] [Google Scholar]
- 34.Maguire, A. J., D. F. J. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Anitmicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marois, C., F. Dufour-Gesbert, and I. Kempf. 2001. Comparison of pulsed-field gel electrophoresis with random amplified polymorphic DNA for typing of Mycoplasma synoviae. Vet. Microbiol. 79:1-9. [DOI] [PubMed] [Google Scholar]
- 36.Marshall, S., C. G. Clark, G. Wang, M. Mulvey, M. T. Kelly, and W. M. Johnson. 1999. Comparison of molecular methods for typing Vibrio parahaemolyticus. J. Clin. Microbiol. 37:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto, Y., F. Ikeda, T. Kamimura, Y. Yokota, and Y. Mine. 1988. Novel plasmid-mediated β-lactamases from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer, J. J., M. D. Lee, C. Lobsinger, T. Brown, M. Maier, and S. G. Thayer. 1998. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis. 42:431-451. [PubMed] [Google Scholar]
- 39.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed.; approved standard. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 40.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed.; approved standard. NCCLS document M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 41.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard. NCCLS document M31-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 42.Navarro, F., E. Perez-Trallero, J. M. Marimon, R. Aliaga, M. Comariz, and B. Mirelis. 2001. CMY-2 producing Salmonella enterica, Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis and Escherichia coli strains isolated in Spain (October 1999-December 2000). J. Antimicrob. Chemother. 48:383-389. [DOI] [PubMed] [Google Scholar]
- 43.Nolan, L. K., R. E. Wooley, J. Brown, J. L. Blue, and M. Camp. 1987. Comparison of virulence factors and antibiotic resistance profiles of Escherichia coli strains from humans and dogs with urinary tract infections. J. Vet. Int. Med. 1:152-157. [DOI] [PubMed] [Google Scholar]
- 44.Normand, E. H., N. R. Gibson, D. J. Taylor, S. Carmichael, and S. W. J. Reid. 2000. Trends of antimicrobial resistance in bacterial isolates from a small animal referral hospital. Vet. Rec. 146:151-155. [DOI] [PubMed] [Google Scholar]
- 45.Olsen, J. E., M. N. Skov, O. Angen, E. J. Threlfall, and M. Bisgaard. 1997. Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype Typhimurium defined by ribotyping, IS200 typing and PFGE. Microbiology 143:1471-1479. [DOI] [PubMed] [Google Scholar]
- 46.Olson, P., A. Hedhammar, A. Faris, K. Krovacek, and T. Wadstrom. 1985. Enterotoxigenic Escherichia coli (ETEC) and Klebsiella pneumoniae isolated from dogs with diarrhoea. Vet. Microbiol. 10:577-589. [DOI] [PubMed] [Google Scholar]
- 47.Pittet, D., D. S. Dharan, S. Touveneau, V. Sauvan, and T. V. Perneger. 1999. Bacterial contamination of the hands of hospital staff during routine patient care. Arch. Intern. Med. 26:821-826. [DOI] [PubMed] [Google Scholar]
- 48.Provence, D. L., and R. Curtiss III. 1994. Gene transfer in gram-negative bacteria, p. 317-347. In P. Gerhardt (ed.), Methods in general and molecular bacteriology. ASM Press, Washington, D.C.
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Samuelsen, O. B. 1998. Efficacy of orally administered florfenicol in the treatment of furunculosis in Atlantic salmon. J. Aquatic Anim. Health 10:56-61. [Google Scholar]
- 51.Schiappa, D. A., M. K. Hayden, M. G. Matushek, F. N. Hashemi, J. Sullivan, K. Y. Smith, D. Miyashiro, J. P. Quinn, R. A. Weinstein, and G. M. Trenholme. 1996. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli bloodstream infection: a case-control and molecular epidemiologic investigation. J. Infect. Dis. 174:529-536. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt, A. S., M. S. Bruun, I. Dalgaard, and J. L. Larsen. 2001. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67:5675-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, D., D. J. Biedenbach, P. L. Winokur, M. A. Pfaller, and P. L. Winokur, M. A. Pfaller, and R. N. Jones. 1999. Phenotypic and genotypic characterizations of Chinese strains of Escherichia coli producing extended-spectrum β-lactamases. Diagn. Microbiol. Infect. Dis. 34:159-164. [DOI] [PubMed] [Google Scholar]
- 54.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 55.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 56.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, R. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thong, K., Y. Ngeow, M. Altwegg, P. Navaratnam, and T. Pang. 1995. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J. Clin. Microbiol. 33:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tillotson, K., C. J. Savage, M. D. Salman, C. R. Gentry-Weeks, D. Rice, P. J. Fedorka-Cray, D. A. Hendrickson, R. L. Jones, A. W. Nelson, and J. L. Traub-Dargatz. 1997. Outbreak of Salmonella infantis infection in a large animal veterinary teaching hospital. J. Am. Vet. Med. Assoc. 211:1554-1557. [PubMed] [Google Scholar]
- 59.Uhaa, I. J., D. W. Hird, D. C. Hirsh, and S. S. Jang. 1988. Case-control study of risk factors associated with nosocomial Salmonella krefeld infection in dogs. Am. J. Vet. Res. 49:1501-1505. [PubMed] [Google Scholar]
- 60.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, D. G., C. R. Hudson, J. J. Maurer, S. Ayers, S. Zhao, M. D. Lee, L. F. Bolton, T. Foley, and J. Sherwood. 2000. Characterization of chloramphenicol and florfenicol resistance in bovine pathogenic Escherichia coli. J. Clin. Microbiol. 38:4593-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, P. A., C. J. McInver, Y. Deng, and W. D. Rawlinson. 2000. Characterization of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, D. J., P. M. Sears, R. N. Gonzalez, B. S. Smith, H. F. Schulte III, G. J. Bennett, H. H. Das, and C. K. Johnson. 1996. Efficacy of florfenicol for treatment of clinical and subclinical bovine mastitis. Am. J. Vet. Res. 57:526-528. [PubMed] [Google Scholar]
- 64.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wittwer, C. T., G. C. Fillmore, and D. R. Hillyard. 1989. Automated polymerase chain reaction capillary tubes with hot air. Nucleic Acids Res. 17:4353-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolated from animals and food. Antimicrob. Agents Chemotherap. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]