Abstract
To characterize the genetic basis of drug resistance in Mycobacterium tuberculosis in Latvia, mutations involved in rifampin (rpoB gene) and isoniazid (katG gene) resistance in DNA from 19 drug-susceptible and 51 multidrug-resistant M. tuberculosis complex isolates were analyzed. The most frequent rpoB gene mutations found by the Line Probe assay were the S531L (14 of 34 isolates), D516V (7 of 34), H526D (4 of 34), and D516Y plus P535S (4 of 34) mutations. Direct sequencing of seven isolates with unclear results from Line Probe assay showed the presence of the L533P mutation and the Q510H plus H526Y (1 of 34) and D516V plus P535S (4 of 34) double mutations, neither of which has been described previously. Single-strand conformation polymorphism analysis showed strand mobility differences between the rifampin-susceptible and -resistant samples for the D516V, H526D, and D516Y plus P535S mutations but not for the S531L mutation. Nucleotide substitution at codon 315 (AGC→ACC) of the katG gene was found in 48 of 51 multidrug-resistant samples by sequencing. Furthermore, katG gene restriction fragment length polymorphism analysis with endonuclease AciI confirmed the nucleotide change in codon 315.
The incidence of tuberculosis (TB) in Latvia, as in the rest of the world, has followed the same increasing trend (3). From 1991 to 1998, the incidence of TB in Latvia increased, reaching a rate of 74 per 100,000 inhabitants. Since then, it has remained at about this level. At the same time, drug resistance among TB patients in Latvia seems to be the highest in the world (12), with primary multidrug resistance found in 8.6% of new cases and in 34.5% of all TB patients in the year 2000. In comparison, the average primary rate of multidrug resistance in Europe as a whole is only 4%. Mutations in the 81-bp hypervariable region of the rpoB gene encoding the RNA polymerase β-subunit are the cause of rifampin (RIF) resistance in 96% of the cases. Among these, missense mutations in codons S531, H526, and D516 are the most frequent (11, 19). Isoniazid (INH) is most often used for TB treatment because of the very high sensitivity of Mycobacterium tuberculosis to this drug (2). In 60 to 70% of the cases, INH resistance is associated with mutations in the catalase-peroxidase-coding katG gene at codon 315 (Ser→ Thr) (7). In this study, we characterized the rpoB and katG gene mutations that are dominant in the Latvian isolates of multidrug-resistant (MDR) M. tuberculosis (16) by commercial Line Probe assay (LiPA), direct sequencing of PCR products, and single-strand conformation polymorphism (SSCP) and PCR-restriction fragment length polymorphism analyses. We suggest that this approach using molecular markers could be useful for predicting multidrug resistance in M. tuberculosis.
Seventy patients from different regions of Latvia admitted to the State Centre for Tuberculosis and Lung Diseases between 1999 and 2000 with clinical symptoms of lung or renal tuberculosis or tuberculous meningitis were included in this study. Diagnosis was confirmed by microscopy and by culturing (18). Patients with a primary infection were preferably selected. They were typical of TB patients in Latvia in that they ranged in age from 25 to 60 years and in that two-thirds of them were male. Cultures of the M. tuberculosis complex were grown on Löwenstein-Jensen medium for 4 to 6 weeks. Drug susceptibility was determined by using the absolute concentration method on slants with the H37Rv strain of M. tuberculosis as the positive control and by using the BACTEC system (6, 13). With the absolute concentration method, resistance was defined as growth on solid media containing graded concentrations of drugs with more than 20 CFU at a specific drug concentration. The breakpoints for INH were 0.2 and 2.0 μg/ml on Löwenstein-Jensen medium and 0.1 μg/ml on the BACTEC system; for RIF, they were 40.0 μg/ml on Löwenstein-Jensen medium and 2.0 μg/ml on the BACTEC system. Of the 70 strains examined, 19 were drug susceptible and 51 were MDR, i.e., resistant to RIF and INH at least. Due to the small amount of DNA from some of the isolates, it was not possible to perform all of the analyses on all 70 isolates. Therefore, only 19 drug-susceptible and 34 MDR M. tuberculosis isolates were analyzed by LiPA. All 51 MDR isolates, however, were analyzed for the katG gene mutations by nucleotide sequencing. High-molecular-weight genomic DNA was isolated from the 70 mycobacterial cultures by the lysozyme/proteinase K cetyltrimethylammonium bromide procedure and precipitated with isopropanol (20). Purified DNA was dissolved in 20 to 50 μl of TE buffer (10 mM Tris HCl-1 mM EDTA [pH 8]). DNA isolated from M. tuberculosis MT14323 and Mycobacterium bovis BCG strains was used for controls.
A commercial INNO-LiPA Rif kit (Innogenetics NV, Ghent, Belgium) was used for the detection of rpoB gene mutations. First, a 256-bp fragment of the gene was amplified with biotinylated primers 71.B (5′-GGTCGGCATGTCGCGGATGG-3′) and 72.B (5′-GCACGTCGCGGACCTCCAGC-3′) flanking the 81-bp region of the rpoB gene (14). Ten to twenty nanograms of DNA diluted in TE buffer was added to the PCR mixture to a final volume of 50 μl [containing PCR buffer with (NH4)2SO4, 2.5 mM MgCl2, 200 μM concentrations of each dNTP, and 1 U of Taq DNA polymerase (Fermentas, Vilnius, Lithuania)]. Ten picomoles of each primer was used for one reaction mixture. The optimized cycling protocol was used in a Progene thermal cycler (Techne, Cambridge, England). PCR products were analyzed in 1.2% agarose gel. In each set of reactions, one negative control and one positive control (from a sensitive strain of M. bovis BCG) were included. Ten microliters of each PCR product was used for the LiPA hybridization, which was performed according to the manufacturer's instructions. Each of the LiPA strips used contained one probe specific for the M. tuberculosis complex, five partially overlapping wild-type probes, and four probes specific for the most common rpoB mutations within the 509- to 534-amino-acid region (D516V, H526Y, H526D, and S531L) (Innogenetics NV). Manual sequencing of the rpoB gene was performed by using a Cycle Reader DNA sequencing kit (Fermentas). Radioactively labeled primers used for manual sequencing were identical to those used for the PCR amplification.
SSCP analysis is based on the ability of single-stranded DNA to undergo a conformational change when a single nucleotide is altered. Such alterations or mutations can then be detected because of the change in DNA mobility in polyacrylamide gel. Here, the 256-bp rpoB fragment was used for SSCP analysis. For better strand separation, we used one biotinylated (reverse) primer, 72.B (17). The biotinylated DNA strand was separated from the unbiotinylated strand by Dynal streptavidin magnetic beads by using the single-stranded DNA purification kit (Labsystems, Helsinki, Finland). In brief, 80 μl of the beads, suspended in 40 μl of binding buffer, was mixed with an equal volume of the PCR product at room temperature for 15 min. After washing the beads with 80 μl of washing buffer, the bound DNA was denatured and incubated for 10 min at room temperature in 80 μl of 0.1 M NaOH. The alkaline solution containing the unbiotinylated strand was aspirated and used for SSCP analysis. The beads with the bound biotinylated strand were washed once with washing buffer and then suspended in 20 μl of concentrating solution. After being heated for 5 min at 96°C with an equal volume of loading buffer (0.05% bromophenol blue, 0.05% xylene cyanol, and 95% formamide), the biotinylated and unbiotinylated DNA samples were snap-cooled and immediately loaded onto the gel. The SSCP analysis of DNA fragments was performed in 6% polyacrylamide gel containing 5% glycerin at 35 mA and 500 V on a 17- by 28-cm gel plate cooled to 5°C (Bio-Rad, Hercules, Calif.). After 8 h of electrophoresis, the separated DNA bands were visualized by silver staining.
A 704-bp fragment of the katG gene (GenBank accession no. X68081) (23) was amplified by PCR using direct primer 74(5′-CGGGATCCGCTGGAGCAGATGGGC-3′), targetingthe 52 terminal nucleotides before codon 315, and reverse primer 75 (5′-CGGAATTCCAGGGTGCGAATGACCT-3′), positioned 158 nucleotides downstream from codon 463 (4). The 50 μl of the PCR mixture that was used contained PCR buffer with (NH4)2SO4, 2.5 mM MgCl2, 200 μM concentrations of each dNTP, and 1 U of Taq DNA polymerase (Fermentas). A 12.5-pmol portion of each primer was used for one reaction mixture. The standard cycling protocol was applied in a Progene thermal cycler (Techne). Amplification products were visualized in 1.5% agarose gel, purified, and used for sequencing and for digestion reactions.
Sequencing of the 704-bp katG fragment was performed with fluorescence-labeled dideoxynucleotide terminators by using an ABI PRISM Big Dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.) and the same primers as those used for the PCR amplification. Nucleotide sequences were analyzed by an ABI PRISM 310 genetic analyzer (Applied Biosystems). PCR was performed on the relevant 704-bp katG fragment, followed by restriction with endonuclease AciI (New England BioLabs, Beverly, Mass.). One INH-sensitive M. tuberculosis sample, BCG, and the MT14323 strain were used as controls. AciI is a rare endonuclease that can recognize 5′-C′CGC-3′ and 5′-GCG′G-3′ sequences and can therefore be used for determining changes in codon 315. Four microliters of PCR product was added to the restriction mixture containing 1 μl of 10× restriction buffer 3 (New England BioLabs), 1 μl of AciI endonuclease (5 U/μl), and 4 μl of H2O. After 2 h of incubation at 37°C, the AciI digestion products were separated in 6% polyacrylamide gel and stained with ethidium bromide.
Nineteen drug-susceptible and 34 MDR DNA isolates were analyzed by the reverse hybridization-based LiPA to determine RIF susceptibility. The remaining 17 RIF-resistant samples from a total of 51 MDR patients were not analyzed by LiPA because of insufficient amounts of DNA.
The LiPA confirmed that the 19 drug-susceptible isolates were RIF susceptible and that the 34 MDR isolates were RIF resistant. The presence of M. tuberculosis complex was confirmed by identification of the oligonucleotide zone specific for this complex. The results obtained with LiPA correlated completely with the in vitro susceptibility results obtained from the culturing. One of the analyzed isolates, however, was a mixture of wild-type and mutant (H526Y) strains. Nine different LiPA patterns were found for the 34 RIF-resistant samples (Table 1). The most frequent rpoB gene mutations were S531L (found in 14 of the 34 isolates), D516V (7 of 34), and H526D (4 of 34). One isolate showed the H526Y mutation in rpoB. Mutations were clearly demonstrated in 27 of the 34 RIF-resistant samples by LiPA. The remaining seven RIF-resistant samples with unclear LiPA patterns were examined further by manual nucleotide sequencing (Table 1). The sequencing results showed the D516V plus P535S double mutation in four of these isolates and the Q510H plus H526Y double mutation in another isolate; these double mutations have not been described previously (11). Altogether, five double mutations of the rpoB gene were found in the 34 MDR isolates examined. Sequencing results for the remaining two (of the 34) samples with unclear but similar LiPA patterns (ΔS5, R4a, and R4b; faint zones) showed the L533P mutation.
TABLE 1.
Mutations in the rpoB gene of 34 RIF-resistant isolates
| INNO-LiPA pattern | Mutation(s) | No. of isolates |
|---|---|---|
| ΔS5, R5, (plus R4aa, R4ba) | S531L | 14 |
| ΔS2, R2 | D516V | 7 |
| ΔS4, R4b | H526D | 4 |
| ΔS2, ΔS5, S4a | D516Y, P535Sb | 3 |
| ΔS2 | D516Y, P535Sb | 1 |
| ΔS5, R4aa, R4ba | L533Pb | 2 |
| R4b | Wild type plus H526D | 1 |
| ΔS1, ΔS4, R4a | Q510H, H526Yb | 1 |
| ΔS4, R4a | H526Y | 1 |
Zone is faint.
Detected only by nucleotide sequencing.
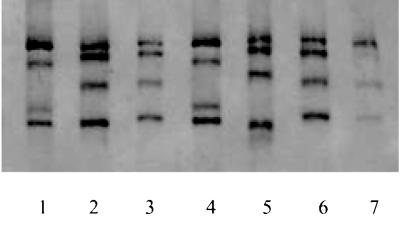
As an alternative to LiPA, the SSCP method was used to examine 23 of the DNA isolates, 8 from drug-susceptible and 15 from RIF-resistant M. tuberculosis cultures. Drug-resistant isolates with the most common rpoB gene mutations in Latvia, including five isolates with the S531L, three isolates with the D516V, two isolates with the H526D, and five isolates with the D516Y plus P5335S substitutions, were selected to evaluate the discriminative power of the SSCP method. Computer modeling (DNA Star) was used for the approximate calculation of the folding of a 256-nucleotide rpoB plus-strand DNA fragment at 37°C for both the wild-type and mutant variants. Three virtually designed folding models of this fragment containing the most frequently observed mutations worldwide, S531L (TCG→TTG), D516V (GAC→GTC), and H526D (CAC→GAC), were analyzed. Double mutations were not included in this modeling. Computer modeling altered the folding of single-stranded DNA in the fragments with the D516V and H526D mutations but not in the fragment with the S531L mutation, where the TCG→TTG change hypothetically should not influence loop interaction (data not shown). The SSCP analysis showed a strand mobility difference between the RIF-susceptible and RIF-resistant samples, except for the five samples with the S531L mutation (Fig. 1). Since S531L is the most common rpoB mutation in our isolates, the employment of the SSCP method for rapid rpoB screening for RIF susceptibility cannot be recommended, at least in Latvia.
FIG. 1.
SSCP analysis of the 256-nucleotide rpoB gene fragments from RIF-resistant M. tuberculosis isolates. Lanes: 1, 3, 4, 5, and 7, rpoB gene fragments with the D516Y plus P535S (lane 1), S531L (lanes 3 and 7), D516V (lane 4), and H526D (lane 5) mutations; 2 and 6, fragments from the RIF-sensitive H37Rv strain of M. tuberculosis.
Fifty-one MDR M. tuberculosis complex isolates showing INH resistance in addition to RIF resistance were analyzed for katG gene mutations. Since mutations in the katG gene have not been observed previously in IHN-susceptible M. tuberculosis, only MDR isolates were included in this study. Automatic sequencing of the 51 INH-resistant samples showed a nucleotide substitution at codon 315 in 48 of the 51 cases, of which 46 cases had a codon change from AGC to ACC and 2 cases had a mixture of AGC changes (to ACC and to ACA). In two other cases, a mixture of AGC and ACC from wild-type and resistant strains was found (Table 2). In all 48 cases, the nucleotide change to ACC or ACA resulted in a change of the amino acid from serine to threonine. In one case, no mutation was found in the analyzed fragment. Initially, two methods, automatic sequencing and AciI restriction analysis, were used to examine the 704-bp katG gene fragment. Later, however, only sequencing, which was found to be more informative and precise for the detection of mutations, was done. Restriction analysis performed with AciI on 35 samples showed a nucleotide change at codon 315 in all samples.
TABLE 2.
Mutations in codon 315 of the katG gene detected by nucleotide sequencing of 51 INH-resistant samples
| Substitution | Amino acid resulting from substitution | No. of samples |
|---|---|---|
| AGC→ACC | Ser→Thr | 46 |
| AGC→ACC and ACA mix | Ser→Thr | 2 |
| AGC→AGC and ACC mix | Ser (wild type) and Thr mix | 2 |
| None | Ser (wild type) | 1 |
The aim of our study was to determine the frequency of mutations underlying drug resistance in M. tuberculosis isolates in Latvia, where a rapid increase in the numbers of TB cases and MDR M. tuberculosis isolates has been seen over the last 10 years. Previously, the most frequently described mutations in the rpoB gene were S531L (47% of isolates) and H526Y (12%) (19). A previous study has shown the presence of the S531L, H526Y, and H526D mutations in 44.5, 10, and 10% of RIF-resistant isolates, respectively (14). Comparison of these results with the results of our study shows that the S531L mutation also predominates in Latvia. In contrast with the results of the previous reports (14, 19), the frequency of the D516V mutation (7 of 34, or 20%) is now higher in Latvia. A high frequency of the D516V mutation (37.9%) has also been described recently in MDR M. tuberculosis isolates from East Hungary (1). Our findings correlate well with this study from a region where less common or novel mutations also occur more frequently. The LiPA was effective in identifying the four most common rpoB mutations: D516V, H526D, H526Y, and S531L. Unfortunately, the method did not identify the types of mutation in the five isolates with double mutations and in the two isolates with the rare L533P mutation, all of which required nucleotide sequencing for identification. Therefore, in a region with a high percentage of double mutations of the rpoB gene, LiPA results may be insufficient. Also, LiPA is relatively expensive (approximately 10 times more expensive than SSCP), and only RIF-resistant isolates were selected for LiPA toward the end of this study.
The high percentage of mutations at position 315 of the katG gene (48 of 51 isolates, or 91%) demonstrates the importance of this codon for the development of INH resistance in M. tuberculosis in Latvia. A high percentage of mutations at codon 315 has also been observed in Russia, where 22 of 24 INH-resistant isolates carried a mutation at codon 315 (Ser→Thr), 1 had a mutation at codon 88 (Gly→Arg), and 1 had a mutation at codon 155 (Tyr→Ser) (10). According to another study, the Arg→Leu mutation at position 463 in the katG gene was found in 30 to 40% of the INH-resistant isolates (15). Further studies of the mutations at codon 315 seem to hold more promise for understanding INH resistance, since the mutation at codon 463 was also found in M. bovis BCG, a strain that is inherently INH sensitive. One MDR isolate showed a lack of mutations in the katG gene fragment analyzed here. According to previous studies, there are more genes responsible for INH resistance, such as the inhA, ahpC, and kasA genes (8, 22) as well as the recently described ndh gene (9). Therefore, mutations responsible for INH resistance in this isolate may be present in any of these genes. The restriction enzyme AciI was used on the 704-kb PCR product and confirmed the high frequency of nucleotide change at codon 315 in the INH-resistant isolates. The AGC→ACC (Ser→Thr) nucleotide change is the mutation most often observed, but other substitutions at codon 315 [AGC→AAC (Ser→Asn), AGC→ATC (Ser→Ile), and AGC→CGC (Ser→Arg)] have been found in some studies (5). The AGC→CGC (Ser→Arg) substitution cannot be distinguished by AciI restriction analysis. Therefore, nucleotide sequencing of the 704-kb katG gene fragment seems to be a more reliable method for detecting the gene responsible for INH resistance. Taking into account the high prevalence of substitutions at codon 315, we propose the use of a shorter katG fragment for the detection of resistance to INH. The high prevalence of the S315 mutation in the katG gene in INH-resistant isolates is clearly associated with multidrug resistance, since all of the isolates with this mutation were also found to be MDR in in vitro susceptibility tests. These data are consistent with the results of a study from The Netherlands where the S315 mutation was found in more than 50% of the INH-resistant isolates (21).
We speculate that one of the reasons for the high level of drug resistance in M. tuberculosis isolates in Latvia may be the selection of strains that develop drug resistance more rapidly. This may explain the high percentage of certain katG gene mutations usually associated with a high level of multidrug resistance in our samples. More-detailed molecular studies with strictly defined patient groups may further elucidate this hypothesis. Molecular markers are a valuable tool for predicting drug resistance in epidemiological studies. At present, unfortunately, most molecular typing and mutation detection methods can only be applied to cultivated bacteria [about 105 copies of the gene(s) are needed to obtain good results in these assays]. Development of methods for uncultivated clinical material will accelerate their introduction in clinical practice throughout the world.
Acknowledgments
This work was supported by grants from the European Community, INCO-Copernicus Project ERBIC15-CT98-0328 (Sven Hoffner, Coordinator), and the Council of Sciences of Latvia (program no. 12-9-1410.
We thank Ilva Pole for the DNA isolation and Liana Pliss for technical assistance. We are also grateful to Petras Stakenas and Daiva Bakonyte (Institute of Biotechnology, Vilnius, Lithuania) for advice and to Eskild Peterson (Statens Serum Institut, Copenhagen, Denmark) and Vitauts Kalnins (University of Toronto, Toronto, Canada) for their help in preparing the manuscript.
REFERENCES
- 1.Bartfai, Z., A. Somoskovi, C. Kodmon, N. Szabo, E. Puskas, L. Kosztolani, E. Farago, J. Mester, L. M. Parsons, and M. Salfinger. 2001. Molecular characterization of Mycobacterium tuberculosis from Hungary by DNA sequencing and the Line Probe assay. J. Clin. Microbiol. 39:3767-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, J., W. A. Lott, B. A. Steinberg, and H. L. Yale. 1952. Chemotherapy of experimental tuberculosis. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am. Rev. Tuberc. 65:357-364. [DOI] [PubMed] [Google Scholar]
- 3.Darzins, E. 1958. The bacteriology of tuberculosis. University of Minnesota Press, Minneapolis.
- 4.Dobner, P., S. Rüsch-Gerdes, G. Bretzel, K. Feldmannn, M. Rifai, T. Löscher, and H. Rinder. 1997. Usefulness of M. tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int. J. Tuberc. Lung Dis. 1:365-369. [PubMed] [Google Scholar]
- 5.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G. Bretzel, V. Sticht-Gron, and H. J. Bremer. 1997. Molecular analysis of katG mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawkins, J., R. Wallace, and B. Brown. 1991. Antibacterial susceptibility tests: mycobacteria, p. 1138-1152. In A. Balows, J. Hausler, K. Herrmann, H. Isenberg, and H. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 7.Heym, B., Y. Zhang, S. Poulet, D. Young, and S. Cole. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for isoniazid susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 13:4255-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, A., I. Lim, L. Lynn, H. Tang, A. Telenti, and S. W. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A. S., A. S. Teo, and S. Y. Wong. 2001. Novel mutation in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45:2157-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martilla, H., H. Soini, E. Eerola, E. Vyshnevskaya, B. Vyshnevskiy, T. Otten, A. Vasilyef, and M. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant M. tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pablos-Mendez, A., M. C. Raviglone, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. B. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, and P. Nunn. 1998. Global surveillance for antituberculosis-drug resistance. N. Engl. J. Med. 338:1641-1649. [DOI] [PubMed] [Google Scholar]
- 13.Roberts, G., E. Koneman, and K. Kim. 1991. Mycobacterium, p. 304-349. In A. Balows, J. Hausler, K. Herrmann, H. Isenberg, and H. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 14.Rossau, R., H. Traore, H. Beenhouwer, W. Mijs, G. Jannes, P. Rijk, and F. Portaels. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouse, D. A., Z. Li, G. H. Bai, and S. L. Morris. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander, P., and E. C. Böttger. 1999. Mycobacteria: genetics of resistance and implications for treatment. Chemotherapy 45:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Selvakumar, N., S. M. Wilson, R. McNerney, and P. R. Narayanan. 1997. Single strand conformation polymorphism profiles with biotinylated PCR products to detect mutations in rpoB gene of Mycobacterium tuberculosis. Curr. Sci. 73:774-777. [Google Scholar]
- 18.Strong, B., and G. Kubica. 1981. Isolation and identification of Mycobacterium tuberculosis. A guide for the level II laboratory. DHHC publication no. (CDC) 81-8390. Centers for Disease Control, Atlanta, Ga.
- 19.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistant mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 20.Van Soolingen, D., P. E. W. Haas, P. W. M. Hermans, and J. D. A. van Embden. 1999. RFLP analysis of mycobacteria. Manual for fingerprinting of M. tuberculosis strains. National Institute of Public Health and Environmental Protection, Bilthoven, The Netherlands.
- 21.Van Soolingen, D., P. E. W. Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, T., and D. Collins. 1996. AhpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025-1034. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, Y., and D. Young. 1994. Strain variation in the katG region of Mycobacterium tuberculosis. Mol. Microbiol. 14:301-308. [DOI] [PubMed] [Google Scholar]