Abstract
Multilocus sequence typing (MLST) of Staphylococcus aureus is well suited to the study of global or long-term epidemiology, but its role in local epidemiology has not been defined. The present study has compared MLST with pulsed-field gel electrophoresis (PFGE) by using S. aureus isolates associated with carriage and disease in a busy regional renal unit. One hundred forty-four patients were prospectively recruited, of whom 103 were receiving hemodialysis and 41 were on continuous ambulatory peritoneal dialysis. Three nasal swab specimens were obtained 1 month apart on entering the study. A nasal swab was positive for S. aureus on at least one occasion in 50 patients (35%). Typing of the 104 carriage isolates demonstrated 21 PFGE types and 21 sequence types (STs). Thirty-one carriers had two or more positive nasal swabs; of these, the isolates in all swabs from a given carrier had identical PFGE types for 29 carriers; the isolates in all of the same 29 swabs had identical STs. The carriage strain in two patients changed both PFGE type and STs during the period of swabbing. Eight patients (6%) had an episode of S. aureus bacteremia during the 12-month study period, and two of these were nasal carriers. One of these invasive isolates had the same PFGE type and ST as the carriage isolate. There were no differences between Simpson's index of diversity for PFGE and Simpson's index of diversity for MLST for both invasive and carriage isolates, suggesting that the two methods have very similar discriminatory abilities. We conclude that PFGE and MLST performed equally in this study.
Molecular typing of the important human pathogen Staphylococcus aureus has been used to examine both long-term or global epidemiology and short-term or local epidemiology. Understanding the genetic structure of the global population over time gives insights into the evolution of bacterial lineages and transmission dynamics and also provides a framework with which to study bacterial pathogenesis. This contrasts with the typing of organisms in a defined setting over a short period of time which is used to study nosocomial outbreaks, local transmission and carriage, and the relationship between isolates associated with carriage and infection in a given individual.
The attributes required of a typing tool vary depending on the application. For example, studies of nosocomial outbreaks caused by strains such as epidemic methicillin-resistant S. aureus strains are commonly performed in many centers; thus, cost, the technical expertise required, speed, reproducibility, and comparability between centers are all important issues. A range of different techniques have been described and evaluated, but pulsed-field gel electrophoresis (PFGE) is the predominant method in use for studies of outbreaks and local epidemiology (2, 5, 8, 14, 17, 20). PFGE has also been successfully applied to isolates separated by geographical location (1), but it is generally considered overdiscriminatory for this type of study since the technique detects genetic variation that accumulates relatively rapidly (10), and even minor genetic changes (for example, a point mutation resulting in creation or loss of a restriction site) can lead to a three-fragment difference in the PFGE gel banding pattern (18). The development of multilocus sequence typing (MLST) represents a major advance in this respect since it relates organisms on the basis of the nucleotide sequences of the fragments of conserved housekeeping genes. MLST of S. aureus was recently described by Enright et al. (9) in a study that examined both methicillin-sensitive and methicillin-resistant strains, and this method has since been used to study the evolution of pandemic clones of methicillin-resistant S. aureus and to identify two ancestral genetic backgrounds (16). However, use of this technique for the study of S. aureus is in its infancy, and its role in the study of local epidemiology is unclear. In principle, the results generated should lead to good reproducibility and to comparability between centers, although this has yet to be formally demonstrated. This contrasts with PFGE, which has been shown to be poorly reproducible between centers (4, 6, 15, 19), leading to considerable effort to introduce standardized protocols to improve reproducibility and technical quality (3, 15). However, MLST is expensive and technically demanding, and it is important to establish whether it represents a clear advantage in terms of discrimination.
The aim of the prospective study described here was to compare MLST with PFGE in a microepidemiological setting. The unit selected for study was a busy regional renal unit, and the questions addressed were those commonly raised in previous studies: to evaluate the discriminatory abilities of the tests when they were applied to isolates associated with carriage and bacteremia, examining carriage and infecting isolates in a given individual and in the patient group as a whole. In view of the potential for PFGE to be more discriminatory than MLST, the study also compared isolate diversity determined on the basis of the results of the two techniques.
MATERIALS AND METHODS
Study design.
This prospective study was conducted on the Oxford Renal Unit at the Churchill Hospital, Oxford, United Kingdom, for 1 year from the beginning of March 1999. The unit serves a population of approximately 2 million and provides care to approximately 290 patients on chronic hemodialysis (HD) and 180 patients on peritoneal dialysis. Approval for the study was obtained from the Central Oxford Research Ethics Committee. All patients with chronic renal failure attending the outpatient HD unit were requested to participate in the study, and informed consent was obtained if they agreed. Those receiving HD for acute renal failure were not included. Patients who were dialyzed via continuous ambulatory peritoneal dialysis (CAPD) were enrolled in the outpatient clinic. Three nasal swab specimens were obtained a minimum of 1 month apart. Episodes of S. aureus bacteremia in these patients were determined (i) between March 1999 and February 2000 and (ii) during the 5 years preceding the study. The demographic and clinical details recorded were age, sex, cause of renal failure, immunosuppression (from drugs, malignancy, or diabetes mellitus), time (months) on dialysis, mode of dialysis, presence of an intravenous device during the study, and death.
Nasal swabs.
A cotton swab was moistened with sterile peptone water saline diluent (PWSD) and rubbed over the anterior nares of both nostrils by using three 360° clockwise movements and three 360° counterclockwise movements. The tip of the swab was broken into a bottle containing 1 ml of PWSD and stored at 4°C until it was returned to the laboratory. The sample was vortexed and diluted 100-fold in PWSD, and 50 μl each of diluted fluid and undiluted fluid was plated onto a mannitol salt agar (MSA) plate, which was incubated at 37°C in air for 48 h. The remainder of the undiluted sample was inoculated into 5 ml of 5% salt broth and incubated at 37°C in air for 5 days. This was subcultured onto MSA plates if it became cloudy during this period or was subcultured on day 5 if the broth remained clear.
Bacterial isolates, isolate identification, and antibiotic susceptibility testing.
Colonies that were isolated from nasal swabs and that were yellow on MSA (mannitol fermenters) were plated to purity on blood agar and incubated at 37°C in air for 24 h. The isolates were identified as S. aureus on the basis of positive catalase, coagulase, and DNase tests. Blood cultures positive for S. aureus during the study period were identified in the Department of Microbiology, John Radcliffe Hospital. Blood cultures were processed in that department by incubation of the bottles for 5 days on the BACTEC 9240 system (Becton Dickinson) and were subcultured according to the standard operating procedures of the laboratory if they were flagged as positive during this period. S. aureus strains isolated from cultures of blood taken from study patients during the previous 5 years were obtained from a freezer archive within the Department of Microbiology. The identities of all blood culture isolates were confirmed to be S. aureus by the methodology described above. Antibiotic susceptibility testing was performed by a comparative disk diffusion method in accordance with British Society for Antimicrobial Chemotherapy guidelines (11). Freezer stocks based on a single colony of S. aureus were stored in Trypticase soy broth with glycerol (15% [vol/vol]) at −80°C.
PFGE.
Chromosomal DNA from S. aureus was prepared in agarose blocks and cleaved with SmaI (New England BioLabs), as described elsewhere (7). The samples were run on a 1% agarose gel (Gibco) in a CHEF DRIII system (Bio-Rad) under the following conditions: run time, 24 h; temperature, 14°C; voltage, 200 V; initial forward time, 1 s; final forward time, 30 s. Bacteriophage lambda concatemers (New England BioLabs) were run in every sixth lane. The gels were stained with ethidium bromide, washed in water, and photographed under UV light by using the Gel Doc 1000 system (Bio-Rad). Two observers analyzed stored images using BioNumerics (version 2.0) software (Applied Maths, Kortrijk, Belgium). PFGE types were defined on the basis of the DNA banding patterns. Isolates with identical patterns were regarded as genotypically indistinguishable, those with one to three band differences were regarded as closely related, those with four to six band differences were regarded as possibly related, and those with seven or more band differences were regarded as unrelated (18). For the primary analysis, organisms were grouped into a single PFGE type if they were indistinguishable or closely related (differing by up to and including three bands).
To assess the effect of the number of band differences used for PFGE type definition on the discriminatory power and utility of PFGE, we conducted two secondary analyses of the PFGE data. In addition to defining PFGE types on the basis of up to and including three band differences, we also reanalyzed the banding patterns grouping isolates with (i) indistinguishable banding patterns (no band differences at all) and (ii) six or fewer band differences. The three criteria examined correspond to the three levels of relatedness discussed by Tenover et al. (18): indistinguishable, closely related, and possibly related. These are based on the putative existence of zero, one, and two genetic differences between any two strains, respectively.
MLST.
Genomic DNA from S. aureus was extracted by using the Wizard Genomic DNA purification kit, with the modification that 30 μg of lysostaphin (Ambi) per ml was added at the cell lysis step. MLST was carried out on all isolates by the methodology described by Enright et al. (9). In brief, fragments of seven unlinked housekeeping genes were amplified by the PCR. The amplified products were precipitated, and both strands were sequenced by using BigDye fluorescent terminators and the primers used in the initial PCR amplification. The sequences obtained were assigned allele numbers following comparison of the DNA sequence with the sequences of previously typed strains by using the MLST website (www.mlst.net). For each isolate, the allele numbers at each of the seven loci defined the allelic profile or sequence type (ST). Both strands of all PCR products were fully sequenced, and novel alleles and STs not found on the MLST website were confirmed by repeating both the PCR and sequencing.
Statistical analysis.
All isolates were typed and taken into account in the analysis of carriage and disease within individuals. However, for analysis of genetic diversity, only carriage or disease isolates which differed by either PFGE or MLST from other isolates of the same source within a given individual were considered. For example, three isolates from three swabs from the same individual identical by PFGE and MLST were considered a single stable carriage isolate. Similarly, blood culture isolates that were obtained during the same clinical episode of sepsis and that were identical PFGE and MLST types were considered the same isolate. Isolate diversity based on PFGE and MLST was assessed by using Simpson's index of diversity, with 95% confidence intervals calculated by the method of Grundmann et al. (12). Proportions were compared between groups by Fisher's exact test, and continuous variables were compared by the Kruskal-Wallis test. Multiple logistic regression analysis was used to determine risk factors for S. aureus carriage and S. aureus bacteremia.
RESULTS
Patient details.
One hundred forty-four patients were recruited into the study, of whom 103 were receiving HD and 41 were on CAPD. The age range was 16 to 88 years (mean age, 59.7 years), and 92 patients (64%) were males. Time on dialysis at the time of recruitment ranged from 1 day to 21 years (median, 33 months; interquartile range, 14 to 55 months). Nine patients (6%) were receiving immunosuppressive drugs. The underlying renal disease was unknown in 37 patients (26%) and was attributed to glomerulonephritis or vasculitis in 33 patients (23%), postrenal causes in 26 patients (18%), diabetic nephropathy in 15 patients (10%), adult polycystic kidney disease in 12 patients (8%), hypertensive nephropathy in 10 patients (7%), pyelonephritis in 7 patients (5%), and miscellaneous other conditions (systemic sclerosis, tuberose sclerosis, myeloma, and drug toxicity) in 4 patients (3%). Eight of the 144 patients (6%) changed the mode of dialysis during the study period, with 7 CAPD patients converting to HD and 1 HD patient transferring to CAPD. By the end of the study year, the cohort was reduced from 144 to 101 (70%); 23 (16%) patients had died, 17 (12%) had undergone successful renal transplants, and 3 (2%) had been transferred to other dialysis units.
S. aureus carriage.
Of the 144 patients, 50 (35%) had a nasal swab positive for S. aureus on at least one occasion during the study (Table 1). There was no difference in carriage rates between CAPD and HD patients (12 of 41 [29%] and 38 of 103 [37%], respectively [P = 0.44]). Due to the deaths and the movement of patients between dialysis units, three consecutive monthly swabs were obtained for 111 of 144 patients, with two swabs being taken from the remainder of the patients. All three swabs were positive for S. aureus in 23 patients (21%), one or two swabs were positive in 19 patients (17%), and S. aureus was not detected in 68 patients (61%). In all, a total of 104 carriage isolates were obtained for study. Age, sex, underlying renal disease (including diabetes), immunosuppressive drugs, time on dialysis, or mode of dialysis had no significant effect on whether a patient was a carrier of S. aureus.
TABLE 1.
MLST sequence types and PFGE genotypes of the nasal carriage and disease isolates evaluated in the present study
| Patient no. | Nasal carriage isolates
|
Disease isolates
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MLSTa ST for swab no.:
|
PFGE genotype for swab no.:
|
MLST ST
|
PFGE genotype
|
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | Retro. | Prosp. | Retro. | Prosp. | |
| 5 | —b | — | — | — | 15 | 37 | ||||
| 9 | 22 | 22 | 23 | 23 | ||||||
| 10 | — | — | — | — | — | — | 45 | 45 | 8 | 8 |
| 13 | 20 | — | 20 | 12 | — | 12 | ||||
| 15 | 15 | 15 | 41 | 41 | ||||||
| 16 | 8 | 8 | 8 | 2 | 2 | 2 | ||||
| 22 | 200 | — | — | 34 | — | — | ||||
| 24 | 49 | 49 | 49 | 10 | 10 | 10 | ||||
| 31 | 30 | 30 | 30 | 13 | 13 | 13 | ||||
| 32 | — | — | — | — | — | — | 188 | 25 | ||
| 33 | 30 | 30 | 211 | 13 | 13 | 7 | ||||
| 34 | 39 | 39 | 39 | 1 | 1 | 1 | ||||
| 36 | 47 | 47 | 47 | 7 | 7 | 7 | 30 | 13 | ||
| 40 | — | — | 213 | — | — | 44 | ||||
| 45 | 25 | 25 | 9 | 4 | 4 | 13 | ||||
| 46 | — | — | 22 | — | — | 23 | ||||
| 47 | — | 25 | — | — | 4 | — | ||||
| 48 | 25 | — | — | 23 | — | — | ||||
| 50 | 30 | — | — | 15 | — | — | 30, 26 | 13, 6 | ||
| 51 | 20 | 20 | 20 | 11 | 11 | 11 | ||||
| 52 | — | — | — | — | — | — | 22 | 19 | ||
| 53 | — | 1 | — | — | 47 | — | ||||
| 54 | — | — | 1 | — | — | 47 | ||||
| 60 | — | — | — | — | — | — | 5 | 21 | ||
| 62 | — | 101 | 101 | — | 5 | 5 | ||||
| 63 | — | — | — | — | — | — | 25 | 6 | ||
| 67 | — | — | — | — | — | — | 189 | 13 | ||
| 69 | 207 | 207 | — | 43 | 43 | — | ||||
| 70 | 12 | — | 13 | — | ||||||
| 73 | — | 15 | — | — | 48 | — | ||||
| 74 | 25 | 6 | ||||||||
| 77 | — | — | — | — | — | — | 18 | 24 | ||
| 82 | — | 22 | — | 23 | ||||||
| 83 | — | — | — | — | 12 | 34 | ||||
| 85 | — | — | 22 | — | — | 6 | 47 | 7 | ||
| 88 | 12 | — | 12 | 34 | — | 34 | ||||
| 89 | 30 | 30 | 30 | 13 | 13 | 13 | 30 | 13 | ||
| 90 | 212 | — | 212 | 45 | — | 45 | ||||
| 92 | 25 | 25 | — | 6 | 6 | — | ||||
| 93 | 1 | 1 | 1 | 5 | 5 | 5 | ||||
| 95 | — | — | — | — | — | — | 26 | 6 | ||
| 97 | — | — | — | — | — | — | 30 | 13 | ||
| 98 | 210 | 210 | 210 | 13 | 13 | 13 | ||||
| 99 | 205 | 47 | ||||||||
| 100 | 30 | 13 | 8 | 18 | ||||||
| 102 | 30 | 30 | 30 | 13 | 13 | 13 | ||||
| 103 | — | — | 30 | 13 | ||||||
| 105 | 1 | — | — | 47 | — | — | 22 | 23 | ||
| 106 | — | — | 45 | 8 | ||||||
| 107 | 30 | 13 | ||||||||
| 110 | — | — | 30 | 13 | ||||||
| 111 | 25 | 25 | 25 | 6 | 6 | 6 | ||||
| 112 | 25 | 25 | 25 | 6 | 6 | 6 | ||||
| 114 | 30 | 30 | 30 | 13 | 13 | 13 | 22 | 35 | ||
| 116 | 25 | 25 | 25 | 6 | 6 | 6 | ||||
| 122 | 39 | — | — | 16 | — | — | 39 | 16 | ||
| 123 | — | — | — | — | — | — | 190 | 20 | ||
| 126 | — | — | 8 | 5 | 2 | 21 | ||||
| 132 | — | 15 | — | — | 41 | — | 45 | 22 | ||
| 133 | 30 | 30 | 30 | 13 | 13 | 13 | ||||
| 137 | 22 | 22 | 22 | 15 | 15 | 15 | ||||
| 139 | 22 | 22 | 22 | 23 | 23 | 23 | ||||
| 140 | 8 | 8 | 8 | 3 | 3 | 3 | ||||
| 141 | 39 | 39 | 39 | 16 | 16 | 16 | ||||
| 142 | 30 | 30 | 30 | 13 | 13 | 13 | ||||
| 144 | 34 | 34 | 34 | 13 | 13 | 13 | ||||
MLST sequence types and PFGE genotypes are shown for all carriage and disease isolates, arranged by patient and time of isolation. PFGE types are from the primary analysis, in which isolates with three or fewer band differences in their PFGE banding patterns were grouped as single types. Data for patients who did not carry S. aureus at any time and for whom no isolates from episodes of bacteremia were available (either during the study year or from the previous 5 years) are not shown. Retro., retrospective; Prosp., prospective.
—, a nasal swab was taken but was negative for S. aureus.
S. aureus bacteremia.
S. aureus bacteremia occurred in 8 of 144 patients (6%) during the prospective study period, all of which were in HD patients in association with the presence of a centrally placed intravenous dialysis line. S. aureus carriage did not predict S. aureus bacteremia, with only two of eight patients (25%) being carriers (Table 1). A search of patient notes and the Microbiology Department database identified a further 23 episodes of S. aureus bacteremia in 22 of the 144 study patients during the 5 years prior to the study period (including 2 patients who went on to have further episodes of sepsis during the study period) (Table 1). The isolates responsible for 20 of these episodes in 19 patients were available from the microbiology freezer file. All but two of these episodes were related to intravenous catheters, and nine (39%) occurred in S. aureus carriers (P was not significant). Neither of the two patients who had both a prospectively and retrospectively documented episode of S. aureus bacteremia was a carrier. Taking together all study patients who had S. aureus bacteremia over the 6-year period (retrospective and prospective), there were no significant associations with age, sex, underlying renal disease, use of immunosuppressive drugs, or time on dialysis.
S. aureus carriage status and S. aureus bacteremia were not significantly associated with fatal outcomes. Thirty-five percent of the 23 patients with fatal cases and 36% of the survivors were carriers on at least one occasion. Two of the eight patients with S. aureus bacteremia died (25%); both deaths were attributable to S. aureus septicemia, representing 9% (95% confidence interval, 1 to 28%) of the deaths in the cohort over the study period.
Methicillin susceptibility and PFGE and MLST types of carriage and blood culture isolates.
A total of 132 isolates (104 carriage isolates and 28 invasive isolates) were available for susceptibility testing and typing. Five isolates from four patients were resistant to methicillin, and all five of these isolates were associated with carriage. There were 31 PFGE restriction types (on the basis of three or fewer band differences) and 28 MLST STs in the same isolate population. Twenty-one MLST STs and 21 PFGE genotypes were detected among the 104 carriage isolates (from 50 patients). The 28 disease isolates (collected over a 6-year period from 25 patients) were more diverse, with 15 STs and 17 PFGE genotypes detected (Table 1). Thirty-one carriers had two or more positive nasal swabs; of these, the isolates in all swabs from a given carrier had identical PFGE types for 29 carriers; the isolates in all of the same 29 swabs had identical MLST STs. In isolates from only two individuals was there a change in ST and PFGE type during the 3-month swabbing period. In no patient was the carriage strain replaced according to the results obtained with one typing system but not the other.
Two of the eight patients with bacteremia were carriers; in one, the invasive isolate had the same PFGE type and MLST type as the carriage isolate. Seven of the 19 patients (37%) with retrospective disease were prospectively defined as carriers, similar to the carriage rate expected by chance. Only two of these episodes were caused by strains related by either MLST or PFGE to those currently carried (for one episode the strains were identical by both MLST and PFGE; for the other episode the strains were identical by MLST only).
Comparison of MLST and PFGE as typing tools.
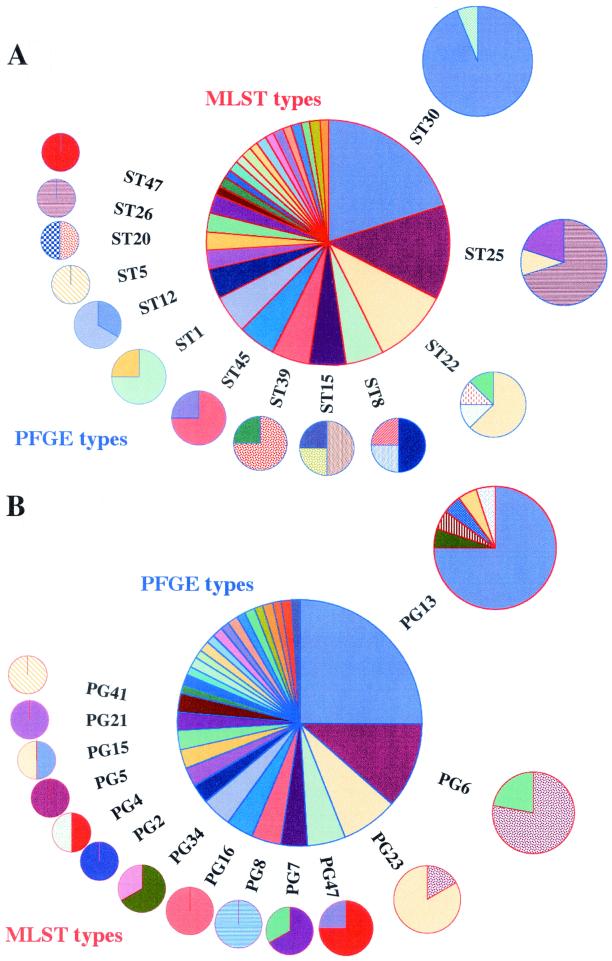
When the same strain was isolated from a given individual on more than one occasion (as defined by genotyping), data for only one isolate were used in subsequent analysis. This gave a total of 52 carriage and 28 disease isolates. The discriminatory powers of MLST and PFGE were compared by examining the number of PFGE types seen among isolates with a particular MLST type and vice versa (Fig. 1). There was no significant difference between the two distributions (P = 0.37). For the 80 invasive and carriage isolates, Simpson's index of diversity for PFGE was 90.5 (95% confidence interval, 87.4 to 95.8), whereas it was 91.6 (95% confidence interval, 89.7 to 95.8) for MLST. The broad overlap in confidence intervals suggests that the two typing methods have very similar discriminatory abilities in this microepidemiological setting.
FIG. 1.
Graphical representation of a comparison of MLST and PFGE as tools for the typing of S. aureus showing the number of PFGE types seen in isolates with a particular MLST type (A) and vice versa (B). PFGE types are from the primary analysis, in which isolates with three or fewer band differences in their PFGE banding patterns were grouped as single types. The size of each pie slice is proportional to the number of isolates with that genotype. Pie slices of the same color and pattern represent identical genotypes within the outer ring of pie charts for each graphic.
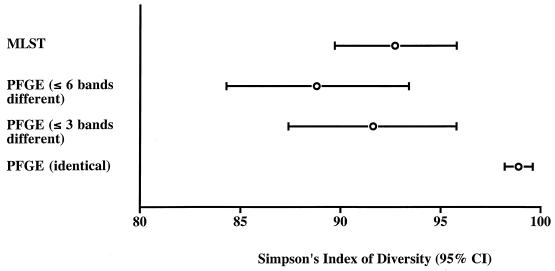
The secondary analyses of the PFGE data revealed 63 PFGE types on the basis of indistinguishable banding patterns and 20 PFGE types on the basis of the grouping of isolates that differed at six or fewer bands (compared with 31 PFGE types on the basis of three or fewer band differences). Simpson's index of diversity was significantly greater for the typing scheme based on indistinguishable banding patterns than for the other two PFGE schemes or MLST, all of which had approximately equivalent discriminatory powers (Fig. 2).
FIG. 2.
Simpson's index of diversity for the isolate collection typed by PFGE (analyzed by using three alternative identity criteria) and by MLST. Error bars represent 95% confidence intervals (CI).
Is there any additional epidemiological utility in the greater discrimination provided by the analysis based on indistinguishable banding patterns? Of the 29 patients with 2 or more nasal swab isolates identical by both MLST and the primary PFGE analysis, 4 individuals had at least one isolate which differed by one or two bands and was therefore given a unique PFGE type under the more discriminatory scheme. Given that the swabs were taken only 1 month apart, it is unlikely that these isolates represent new colonizing strains that happened to be very similar by PFGE and identical by MLST.
Genetic diversity in carriage and disease.
The most common PFGE type in the prospective study, type 13, was equally prevalent in both the carriage and the invasive groups of isolates (13 [26%] versus 7 [25%], respectively). Apart from this one PFGE type, there was little overlap in PFGE types between the carriage, prospective invasive, and retrospective invasive groups. No other type was found in all three groups, and 22 of the 31 PFGE types were unique to a single clinical group. However, there was no evidence of a preponderance of a particular PFGE type in either the disease or the carriage isolate population. The MLST results were similar; two STs (STs 30 and 22) were isolated from all three clinical groups, and there was no significant difference in the distribution of STs between the clinical groups.
DISCUSSION
MLST of S. aureus assesses variations in housekeeping genes that accumulate slowly in the population and that are assumed to be selectively neutral (9). It is likely to prove to be an invaluable research technique for the study of the global epidemiology of S. aureus and has the added advantage that sequence data can be stored in a central database, providing an ever-expanding resource. However, given its theoretical potential for good reproducibility both within and between laboratories, studies are now required to examine the role of this tool in the study of local epidemiology both in the research setting and more generally in clinical practice, for example, during the investigation and management of outbreaks.
The design of this study was chosen to reflect one of the scenarios in which typing is used, that is, for examination of nasal carriage and episodes of bacteremia in individuals over the short term by comparing strains in a given individual and in the group as a whole. The renal unit setting was used because the prevalence of S. aureus carriage and the incidence of S. aureus bacteremia are reported in the published literature to be high (13, 21, 23). Our findings indicated that the levels of discrimination by MLST and PFGE were comparable if isolates are grouped into PFGE types on the basis of three or fewer band differences (corresponding to the closely related category in the criteria of Tenover et al. [18]). By use of this criterion, MLST and PFGE performed equally in demonstrating the relatedness of strains associated with nasal carriage in a given individual and the relatedness between a carriage strain and a prospectively isolated invasive strain in one patient. Small variations in the PFGE banding patterns of carriage isolates from the same individual suggest that for microepidemiological purposes the treatment of unique banding patterns as unique PFGE types leads to overdiscrimination. Conversely, there was little practical difference between three or fewer band differences and six or fewer band differences by PFGE as the definition of identity.
The comparison between disease and carriage isolates within individuals was limited by the finding that only two of the eight prospectively identified cases of bacteremia occurred in carriers. Previous studies have identified S. aureus carriage as a risk factor for bacteremia (22, 23), but S. aureus carriage did not predict bacteremia in our study. Given the relatively short duration over which isolates were prospectively collected, one of the rationales for the inclusion of isolates associated with bacteremia from the 5-year period prior to the study was to include a potentially more genetically diverse bacterial population from the same setting. This theoretically represents a more stringent test for PFGE than that posed by a 1-year sampling time frame, potentially resulting in a higher index of diversity for PFGE than for MLST. However, in the primary analysis the index of diversity was comparable between the two techniques.
Given our findings, and extrapolating to surmise that the findings would be mirrored if the techniques were compared over briefer periods of time (such as during an outbreak), what is the role of MLST in studies other than those evaluating long-term or global epidemiology? One possible application is the comparison of isolates collected over a short period in different units or geographical locations. Identical strains may be more easily identified by MLST, given the potential for the reproducibility of results between units and the portability and unequivocal nature of sequence data. This could provide an exciting resource and represents a clear advantage over PFGE. However, the reproducibility of MLST results between centers cannot be assumed and requires further study. Errors can arise with MLST at a number of levels. First, MLST is intolerant of any errors in sequencing; a single nucleotide error may result in the designation of a new allele and a new ST. Second, unlike PFGE, MLST involves collation of the results of seven separate experiments for each organism, and hence, any pipetting, sequencer gel tracking, or other sequencing errors can lead to the generation of new recombinant STs. These sources of potential error pose a major challenge for the curators of MLST database websites. However, the centralization of MLST sequence and allele profile data could prove to be a major strength of MLST, as with appropriate quality control protocols in place, curators can both ensure the integrity of alleles and STs and provide a valuable open resource for the placement of a typed isolate in its global epidemiological context.
When deciding on a typing system for a particular epidemiological need, careful consideration should be given to the time, cost, and expertise required to perform MLST compared with those required to perform PFGE. For many strictly epidemiological applications, PFGE will remain the typing system of choice, with MLST reserved for long-term epidemiological and population genetic studies. This balance may well change with the inevitable rapid advances and improvements in sequencing technology and its automation.
Acknowledgments
Funding was provided by the Oxford Kidney Unit Trust Fund and by the Wellcome Trust.
We thank the medical and nursing staff of the Oxford Renal Unit for assistance during patient recruitment into the study and collection of bacterial isolates. We also thank F. Cassidy for assistance with sample collection and isolate identification and Mark Enright and Brian Spratt for invaluable help with setting up MLST.
REFERENCES
- 1.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, H. R., J. D. Lian, K. H. Shu, C. H. Cheng, M. J. Wu, C. H. Chen, Y. J. Lau, and B. S. Hu. 2000. Use of pulsed-field gel electrophoresis in the analysis of recurrent Staphylococcus aureus infections in patients on continuous ambulatory peritoneal dialysis. Am. J. Nephrol. 20:463-467. [DOI] [PubMed] [Google Scholar]
- 3.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aries de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 5.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 6.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and the European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Silva, G. D., A. Justice, A. R. Wilkinson, J. Buttery, M. Herbert, N. P. Day, and S. J. Peacock. 2001. Genetic population structure of coagulase-negative staphylococci associated with carriage and disease in preterm infants. Clin. Infect. Dis. 33:1520-1528. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez, M. A., H. de Lencastre, J. Linares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 11.Gosden, P. E., J. M. Andrews, K. E. Bowker, H. A. Holt, A. P. MacGowan, D. S. Reeves, J. Sunderland, and R. Wise. 1998. Comparison of the modified Stokes' method of susceptibility testing with results obtained using MIC methods and British Society of Antimicrobial Chemotherapy breakpoints. J. Antimicrob. Chemother. 42:161-169. [DOI] [PubMed] [Google Scholar]
- 12.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirmani, N., C. U. Tuazon, H. W. Murray, A. E. Parrish, and J. N. Sheagren. 1978. Staphylococcus aureus carriage rate of patients receiving long-term hemodialysis. Arch. Intern. Med. 138:1657-1659. [PubMed] [Google Scholar]
- 14.Monsen, T., C. Olofsson, M. Ronnmark, and J. Wistrom. 2000. Clonal spread of staphylococci among patients with peritonitis associated with continuous ambulatory peritoneal dialysis. Kidney Int. 57:613-618. [DOI] [PubMed] [Google Scholar]
- 15.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, and M. Alfa. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 17.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Eiff, C., K. Becker, K. Machka, H. Stammer, G. Peters, et al. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 21.Wanten, G. J., P. van Oost, P. M. Schneeberger, and M. I. Koolen. 1996. Nasal carriage and peritonitis by Staphylococcus aureus in patients on continuous ambulatory peritoneal dialysis: a prospective study. Periton. Dial. Int. 16:352-356. [PubMed] [Google Scholar]
- 22.Yu, V. L., A. Goetz, M. Wagener, P. B. Smith, J. D. Rihs, J. Hanchett, and J. J. Zuravleff. 1986. Staphylococcus aureus nasal carriage and infection in patients on hemodialysis. Efficacy of antibiotic prophylaxis. N. Engl. J. Med. 315:91-96. [DOI] [PubMed] [Google Scholar]
- 23.Zimakoff, J., F. Bangsgaard Pedersen, L. Bergen, J. Baago-Nielsen, B. Daldorph, F. Espersen, B. Gahrn Hansen, N. Hoiby, O. B. Jepsen, P. Joffe, H. J. Kolmos, M. Klausen, K. Kristoffersen, J. Ladefoged, S. Olesen-Larsen, V. T. Rosdahl, J. Scheibel, B. Storm, P. Tofte-Jensen, et al. 1996. Staphylococcus aureus carriage and infections among patients in four haemo- and peritoneal-dialysis centres in Denmark. J. Hosp. Infect. 33:289-300. [DOI] [PubMed] [Google Scholar]