Abstract
Cell proliferation may be altered in many diseases, but it is uncertain exactly how to measure total numbers of divisions. Although it is impossible to count every division directly, potentially total numbers of stem cell divisions since birth may be inferred from numbers of somatic errors. The idea is that divisions are surreptitiously recorded by random errors that occur during replication. To test this “molecular clock” hypothesis, epigenetic errors encoded in certain methylation patterns were counted in glands from 30 uteri. Endometrial divisions can differ among women because of differences in estrogen exposures or numbers of menstrual cycles. Consistent with an association between mitotic age and methylation, there was an age-related increase in methylation with stable levels after menopause, and significantly less methylation was observed in lean or older multiparous women. Methylation patterns were diverse and more consistent with niche rather than immortal stem cell lineages. There was no evidence for decreased stem cell survival with aging. An ability to count lifetime numbers of stem cell divisions covertly recorded by random replication errors provides new opportunities to link cell proliferation with aging and cancer.
Keywords: cancer, endometrium, molecular clock
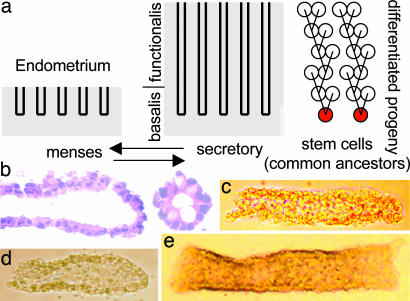
Endometrial divisions are modulated by estrogen (1), with rapid regeneration after menses, followed by maturation and loss in the absence of implantation. Estrogen is thought to stimulate cell proliferation, resulting in higher risks for carcinoma. For example, nulliparous women (those with no live births) have more menstrual cycles and are at higher risk for cancer compared with multiparous women. However, the biology linking cell proliferation to cancer has been difficult to study, because it is impossible to count directly lifetime numbers of divisions. Higher mitotic indices are associated with elevated estrogen levels (1), but these measurements may primarily reflect the proliferation of differentiated cells that are shed with menstruation, instead of stem cells that persist long enough to accumulate the multiple mutations required for transformation. Endometrial glands, like all epithelial tissues (2), are thought to contain small numbers of stem cells that allow for self renewal, multilineage growth, differentiation, and degeneration (3). Very little is known about human endometrial stem cells (4), but they persist through menstruation and therefore are expected to reside in the basalis (Fig. 1a).
Fig. 1.
Endometrial glands. (a) The superficial endometrium or functionalis has growth and degeneration cycles. Stem cells are located in the basalis and are common ancestors or progenitors of differentiated cells within a gland. (b) Endometrial gland fragments isolated free of underlying stroma (hematoxylin/eosin). (c) Thin gland tubes (patient 3). (d) Glands with closed ends (patient 10). (e) Wide gland tubes (patient 3).
Epithelial stem cells have been difficult to characterize, because specific markers are not available. Unknown are the numbers of stems cells per gland, how often they divide, and whether their numbers decline with age. Stem cells are usually identified by an ability to repopulate tissues [such as for hematopoietic stem cells (5)] or through fate-marking studies in experimental animals (6). Such assays are currently impractical for most human epithelia.
Given the impracticality of introducing experimental fate markers into humans, an alternative is the reconstruction of somatic cell lineages from random replication errors. The idea is that all cells are related to each other through a single somatic cell tree that originates from the zygote, and cell divisions and ancestry may be surreptitiously recorded within genomes by replication errors that naturally accumulate in a clock-like manner during aging. Adult stem cells can be defined as progenitors or common (“stem”) ancestors of differentiated cells within clonal units such as single endometrial glands (Fig. 1a).
In theory, the past of any biologic lineage can be reconstructed from present-day sequences. However, such molecular clock approaches are controversial, because they use quantitative models that depend on a number of unverified assumptions (7). For example, replication errors are stochastic, and it is uncertain whether average error rates are constant, because many factors may potentially change error rates. Nevertheless, sequences are commonly used to infer the past, because they appear empirically to reconstruct accurate ancestral trees (7). Sequence errors are too rare to document somatic cell genealogies, but somatic epigenetic errors are also inherited, and methylation measurably increases with age in certain CpG-rich sequences (8, 9). By analogy, although some methylation may result from processes unrelated to numbers of divisions since birth, it is still possible to test a somatic cell epigenetic molecular clock with relatively well characterized tissues. Women of the same chronological age may have different estrogen exposures; thus endometrium provides an opportunity to test whether their methylation patterns represent random replication errors that have accumulated since birth in a clock-like manner. Multiparous women should have fewer menstrual cycles and therefore less methylation than women with fewer children. Obese women should accumulate more methylation from more cell divisions stimulated by higher estrogen levels (1).
Methods
The essential idea is that certain sequences or “molecular clocks” are unmethylated at birth and become progressively more methylated from random errors that may occur with cell division. Therefore, more errors or methylation should be observed in cells with greater numbers of divisions since birth. This hypothesis was tested with endometrial glands, because different numbers of divisions are expected for women of different ages, parity, or body habitus.
Gland Isolation. Fresh uteri were obtained from 30 women undergoing hysterectomy for a variety of diseases (Table 1). Clinical information was obtained from their medical records. Although hormone therapy influences endometrial divisions (1), and some of the women received exogenous estrogen, this information was not uniformly documented and therefore not analyzed in this study. Glands free of stroma were isolated with a procedure originally developed for the colon (10). Grossly normal endometrium and underlying myometrium (≈1 cm2) were sampled from the fundus, sectioned into 0.2- to 0.3-cm cubes, and incubated in 50 ml of Hanks' solution with 30 mM EDTA for 45 min at room temperature. For 15 or more additional minutes, a stir bar was used for vigorous agitation to help release the glands (Fig. 1 b-e). Glands from postmenopausal women were relatively small and few in number, but only eight glands per uterus were needed for analysis.
Table 1. Patients.
| Patient | Age, years | Methylation, % | Parity | BMI | Menopause | Surgery |
|---|---|---|---|---|---|---|
| 1 | 17 | 15.0 | G090 | 31.0 | No | Ovarian CA |
| 2 | 32 | 27.1 | G2P2 | 24.5 | No | Cervical CA |
| 3 | 33 | 15.8 | G5P5 | 27.3 | No | Cervical CA |
| 4 | 36 | 30.9 | G7P5 | 48.2 | No | Dysmenorrhea |
| 5 | 38 | 35.0 | G0P0 | 24.7 | No | Rectal CA |
| 6 | 40 | 43.8 | G4P4 | 31.7 | No | Myoma |
| 7 | 42 | 43.4 | G2P2 | 21.5 | No | Myoma |
| 8 | 43 | 23.4 | G3P0 | 24.4 | No | Dysmenorrhea |
| 9 | 43 | 46.9 | G2P0 | 25.9 | Yes | Ovarian CA |
| 10 | 46 | 35.5 | G0P0 | 19.4 | No | Dysmenorrhea |
| 11 | 46 | 50.8 | G1P1 | 21.2 | No | Ovarian CA |
| 12 | 46 | 43.8 | G10P7 | 30.5 | No | Prolapse |
| 13 | 53 | 53.1 | G3P2 | 40.6 | Yes | Myoma |
| 14 | 54 | 58.4 | G2P2 | 26.3 | No | Ovarian CA |
| 15 | 55 | 47.9 | G8P8 | 25.6 | Yes | Cervical CA |
| 16 | 56 | 47.0 | G10P7 | 23.7 | No | Prolapse |
| 17 | 58 | 74.0 | G1P1 | 30.3 | Yes | Dermoid Cyst |
| 18 | 59 | 53.1 | G3P2 | 22.1 | Yes | Myoma |
| 19 | 64 | 42.4 | G4P4 | 19.4 | Yes | Sarcoma |
| 20 | 65 | 46.5 | G4P4 | 27.7 | Yes | Ovarian CA |
| 21 | 65 | 53.9 | G8P2 | 37.0 | Yes | Sarcoma |
| 22 | 66 | 66.4 | G0P0 | 37.4 | Yes | Bladder CA |
| 23 | 66 | 43.8 | G4P4 | 25.5 | Yes | Cervical CA |
| 24 | 70 | 28.5 | G4P4 | 22.7 | Yes | Bladder CA |
| 25 | 72 | 38.7 | G2P2 | 41.2 | Yes | Myoma |
| 26 | 78 | 71.5 | G3P3 | 24.7 | Yes | Bladder CA |
| 27 | 81 | 52.3 | G2P2 | 34.6 | Yes | Bladder CA |
| 28 | 81 | 38.3 | G4P4 | 15.6 | Yes | Bladder CA |
| 29 | 85 | 44.5 | G12P11 | 20.4 | Yes | Cervical CA |
| 30 | 87 | 51.6 | G2P2 | 30.1 | Yes | Bladder CA |
G, gravida (times pregnant); P, parity (live births); CA, carcinoma.
Individual gland fragments were identified under a dissecting microscope and placed into 500-μl microfuge tubes by using a pipetman. DNA was extracted with a 10-μl solution (100 mM Tris·HCl/4 mM EDTA, pH 8.0/200 μg/ml proteinase K) for 2 hr at 56°C, followed by boiling for 5 min. Cytosine bases were converted by bisulfite treatment by using an agarose bead method (9). Agarose bead sizes were 20-30 μl.
CpG Methylation Analysis. Bisulfite-converted DNA was amplified by PCR for 42 cycles at CSX (eight CpG sites) or CSX6 (six CpG sites and a single-nucleotide polymorphism) with primers illustrated in Fig. 2. The CSX gene on chromosome 5q34 is expressed only in the developing heart (9), and therefore its methylation is unlikely to confer selection in the endometrium. Each gland was amplified in duplicate by using 2-3 μl of the agarose beads, and the PCR products were mixed before cloning (TOPO TA Cloning kit, Invitrogen). Eight clones were sequenced from each gland, and eight glands were analyzed from each uterus. Clones with incomplete bisulfite conversion (Cs at non-CpG sites) were discarded from the analysis.
Fig. 2.
Typical CSX methylation patterns. Each tag is arranged in a 5′ to 3′ horizontal order, with open circles representing unmethylated CpG sites and filled circles representing methylated sites. Eight tags are sampled from each gland. There are eight glands and a total of 64 tags per uterus. Tags are different within and between glands in the same uterus. Below is the 3′ region of the CSX gene, illustrating primer locations (underlined) and CpG sites in the CSX tag (red) and CSX6 tag (yellow). The CSX6 tag has a G/T polymorphism that distinguishes alleles in heterozygous individuals.
Endometrial Gland Modeling. Each sequenced CSX clone or “tag” has 1 of 256 (i.e., 28) possible unique patterns (5′ to 3′ order of methylation) and a percent methylation. Each gland (eight tags) is summarized by average tag percent methylation and gland diversity (number of unique tags among the eight sampled tags). Each uterus (64 tags) is summarized by average gland methylation and diversity.
The goal was to find gland scenarios consistent with endometrial biology and the experimental data. Our model is essentially the same as previously used to describe colon crypts (9), modified to accommodate larger endometrial glands (≈12,000 cells) and variations in division rates during a woman's life. Stem cell lineages may be immortal, or their survival may be probabilistic, as with niches. Immortal stem cell numbers are constant, because they always produce a single stem cell daughter. Niche stem cell numbers are also constant, but stem cells may yield either zero, one, or two stem cell daughters. Therefore, niche stem cell lineages may also become extinct or may expand.
Individual endometrial glands were simulated starting with ≈12,000 cells and a given number (n) of stem cells. Variables are defined in Table 4, which is published as supporting information on the PNAS web site, and different stem cell scenarios are defined in Table 5, which is published as supporting information on the PNAS web site. The variables include numbers of stem cells per gland (4-512); how often they divide (once per week to 1.25 times per day); how often they produce one, two, or zero stem cell daughters; and methylation (2-4.5 × 10-5 per CpG site per division) and dimethylation (5 × 10-6 per CpG site per division) error rates. We emphasize that parameter values are chosen to be consistent with endometrial biology, and further experiments are needed to better define these values. Stem cell division may yield either zero, one, or two stem cell daughters with respective probabilities P0, P1, and P2, with P0+P1+P2 = 1 and P0 = P2. Because total stem cell numbers remain fixed at n, stem cells do not divide independently of each other (see ref. 9; see also Supporting Text, which is published as supporting information on the PNAS web site). With immortal stem cell lineages, P1 = 1, and with niches, P1 < 1. To produce ≈12,000 cells per gland, nonstem cell daughters may also divide a number of times (D), but they always produce two nonstem cell daughters. The lifespan of nonstem cell daughters is limited, because they sequentially die to maintain a constant number of gland cells.
Methylation patterns or tags are replicated with cell division with probability Pm of methylating an unmethylated CpG site. Demethylation is also possible with a probability of Pd. Errors are independent between CpG sites, and all tags start at birth unmethylated [consistent with observations that most CpG islands are unmethylated at birth (11)]. We use two tags, CSX with eight CpG sites and CSX6 with six CpG sites.
Endometrial cell division rates are modeled as slow before menarche (R1), fast during menstruation (R2, ages 12-52 years), and slow after menopause (R3). Mimicking the experimental approach, eight tags are randomly sampled from the gland after a given number of divisions. Percent methylation, gland diversity (numbers of unique tags per eight sampled tags), and intragland tag distance (average Hamming distance) are calculated for each simulated gland.
The size of an endometrial gland varies through life. This potential complication was not included in our model, because simulated outcomes are essentially identical, whether analyzing small (2,000 cells) or larger (24,000 cells) glands (data not shown). This reflects the fact that methylation patterns primarily reflect stem cell divisions, because errors in nonstem cells cannot accumulate. Endometrial glands were simulated (1,000 trials) to yield average values and intervals that include 95% of simulated values.
Results
A somatic cell “molecular clock” hypothesis postulates that numbers of divisions since birth (mitotic age) may be inferred by counting somatic errors. Application requires the isolation of specific cell populations from individuals of different ages, the measurement of multiple sequences (because errors are stochastic), and a quantitative analysis to interpret the genealogic information encoded by random replication errors. The endometrium is composed of multiple cell types, and the first step was the isolation of gland fragments from 30 fresh uteri (Table 1). Microscopic examination revealed primarily an epithelial composition (Fig. 1 b-e). The glands appeared normal, but the cells have divided many times under normal and possibly aberrant hormonal influences, which may be present in some of the eight patients with dysmenorrhea or ovarian cancer (Table 1). Regardless of why cells have divided, in theory, divisions should be recorded within their genomes by random replication errors, with numbers of errors proportional to total numbers of divisions. We chose to count epigenetic errors, because certain CpG rich sequences are unmethylated at birth and measurably accumulate methylation with age (8, 9). DNA was extracted from individual glands, bisulfite-treated, and amplified at our molecular clock, which is a CpG-rich locus (CSX) containing eight CpG sites (Fig. 2). The PCR products were cloned into bacteria and individual clones sequenced. To generate sufficient data for a molecular clock analysis, eight glands, each with eight sequences, were analyzed from each uterus. Consistent with random replication errors, methylation patterns (5′ to 3′ order of methylation) or “tags” were diverse between and within glands from the same uterus (Fig. 2).
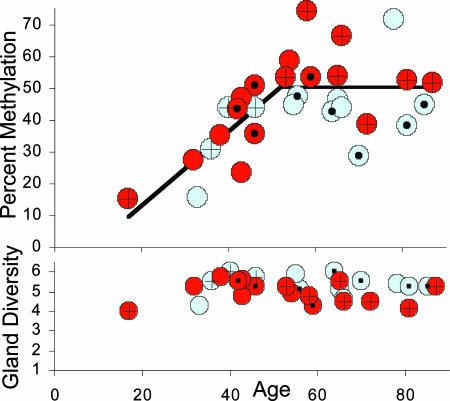
Although individual gland values were scattered, consistent with a clock hypothesis, average uterine values revealed an age-related increase in methylation and relatively constant gland diversity (Fig. 3). Assuming a link between somatic errors and mitotic ages, methylation should primarily increase during menstruation (ages 12-52 years). A regression line indicates that methylation increases after menarche with relatively stable levels after 52 years of age (Fig. 3).
Fig. 3.
Average uterine percent methylation increased during menstruation (ages 12-52 years) and was relatively stable after menopause. A regression analysis of these data was consistent with two lines (as opposed to one; F1,27 = 10.55, P = 0.003), with methylation increasing up to age 52 years and no increase afterward (F1,27 = 0.33, P = 0.57). Red circles indicate two or fewer children; blue circles, three or more children; plus marks indicate obesity (BMI >30) and solid circles lean women (BMI <24). Average uterine gland diversity (numbers of unique tags among eight sampled tags) was stable with age. More complete graphs that include individual gland values are illustrated in Fig. 7, which is published as supporting information on the PNAS web site.
To further distinguish between mitotic and chronological ages, women were divided by parity and body habitus, because endometrial divisions are decreased by parity and increased by obesity (1). Consistent with a link between cell division and methylation, women with fewer children or obesity [body mass index (BMI) >30] generally had greater methylation (Fig. 3). For a statistical analysis, only women >52 years old were compared, because cell divisions are so age-dependent before menopause. Significantly less methylation was observed in multiparous women with three or more children and lean (BMI <24) vs. obese women (Table 2). Gland diversity was also greater in lean and multiparous women, although significant only for multiparous women.
Table 2. Parity and obesity (women >52 years old).
| Parity
|
Obesity
|
|||||
|---|---|---|---|---|---|---|
| <3 (n = 9) | >2 (n = 9) | P value* | BMI >30 (n = 7) | BMI <24 (n = 6) | P value | |
| Average | 1.67 | 5.44 | 35.9 | 20.7 | ||
| Average age, years | 66.1 ± 4.0† | 68.9 ± 3.5 | 0.30 | 68.9 ± 4.6 | 69.2 ± 4.8 | 0.48 |
| Percent methylation | 55.7 ± 3.3 | 45.3 ± 3.8 | 0.027 | 55.7 ± 4.3 | 42.4 ± 3.4 | 0.017 |
| Gland diversity | 4.78 ± 0.16 | 5.35 ± 0.14 | 0.008 | 4.84 ± 0.19 | 5.23 ± 0.23 | 0.11 |
One-sided t test.
Standard error of the mean.
To more fully extract the ancestry recorded by seemingly random tag patterns, a quantitative analysis using constant error rates and different stem cell dynamics was applied. Immortal lineages in which a stem cell always divides asymmetrically to produce one stem cell and one differentiated daughter were inconsistent with the methylation patterns (see Fig. 8 and Table 6, which are published as supporting information on the PNAS web site, for details). A more consistent model is a stem cell niche in which stem cell numbers rather than lineages remain constant (Fig. 4). A small number of stem cells in the basalis divide to reproduce themselves and progeny that leave the niche and differentiate. Niche stem cell division is predominantly asymmetric, but rarely will a stem cell produce two stem cell daughters balanced by another stem cell that produces two differentiated daughters. The exact size of the endometrial niche cannot be determined from the data (Fig. 4), but if the niche contains 64 stem cells, asymmetric stem cell division occurs ≈98% of the time. With these niche dynamics, random stem cell loss with replacement repeatedly leads to the loss of all stem cell lineages except one, estimated (see Supporting Text for calculations) to recur after a median interval of ≈10,000 divisions (95% range ≈ 4,000-28,000 divisions).
Fig. 4.
Stem cell niche. Stem cells are defined by their location within the niche, and their numbers are constant. After division, cells that leave the niche differentiate and eventually die. Niche stem cell lineages may either expand or become extinct, because stem cell division may be asymmetric (one stem and one differentiated daughter) or symmetric (two stem or two differentiated daughter cells). The exact size of the niche and extent of symmetric divisions are uncertain, because a variety of combinations are consistent with the experimental data (see table). A large niche with frequent symmetric division is equivalent to a smaller niche with less frequent symmetric division. Eventually all current-day niche stem cell lineages are lost except one, which recurs with a median interval of ≈10,000 divisions. Below are CSX endometrial gland simulations that assume 64 stem cells, P1 of 0.98, a methylation error rate of 4.5 × 10-5, and a demethylation error rate of 5 × 10-6 per CpG site per division. Smaller or larger niches as in the table have similar outcomes (see Supporting Text). The gray line simulates nulliparous women (no live births) with one division per week before menarche, one division per day between ages 12 and 52, and one division per week after menopause. Multiparity (blue line) is simulated with 0.75 divisions per day between menarche and menopause. Obesity (red line) is simulated with 1.25 divisions per day after menarche. Dotted lines include 95% of simulated gland outcomes indicating the scatter expected from stochastic errors and stem cell turnover.
Stem cell division is modeled (Fig. 4) as minimal before menarche (once per week), rapid with menstruation (average of one division per day), and minimal after menopause (once per week). Ages at menarche or menopause are variable among women but are modeled as occurring at 12 and 52 years of age. Although every gland within a uterus follows the same model, consistent with the diverse methylation patterns sampled from each uterus (Fig. 2), each gland may become different, because stem cell turnover and errors are stochastic. For example, in a 67-year-old nulliparous woman, 95% of simulated gland outcomes are between 26% and 70% methylation and three to seven unique tags per gland. Multiparous women may be simulated by assuming that pregnancy halts stem cell division, with an average of 0.75 stem cell divisions per day (vs. one division per day in nulliparous women) before menopause (Fig. 4). Obesity may be simulated by an increase to 1.25 stem cell divisions per day after menses. These stem cell mitotic rate changes generate lower average percent methylation and greater gland diversity in lean or multiparous postmenopausal women, although stochastic errors and stem cell turnover can mask these trends in individual uteri (Fig. 4). Consistent with these expectations, average methylation was significantly lower and average gland diversity higher in lean or multiparous women children (Table 2).
Different methylation patterns within the same uterus (Fig. 2) are consistent with individual glands maintained by distinct stem cell niches that evolve independently. However, it is uncertain whether different parts of a gland may divide and accumulate methylation at different rates. We searched for such spatial heterogeneity by comparing gland fragments with small or large diameters or closed ends (Fig. 1 c-e). There were no significant differences in percent methylation or tag diversity between different-shaped glands compared among uteri (Table 3). This lack of heterogeneity is consistent with glands composed of related cells separated from each other and their stem cells by relatively few divisions.
Table 3. Comparisons among glands of different shapes.
| Age | Percent methylation | Gland diversity | |
|---|---|---|---|
| Premenopausal women (n = 12) | |||
| Closed ends* (n = 31) | 42.5 | 37.3 ± 4.0† | 5.23 ± 0.30 |
| Thin tubes (n = 46) | 42.7 | 38.7 ± 2.9 | 5.48 ± 0.19 |
| Wide tubes (n = 18) | 42.1 | 37.2 ± 4.8 | 4.94 ± 0.39 |
| Postmenopausal women (n = 15) | |||
| Closed ends (n = 39) | 67.1 | 55.0 ± 3.7 | 4.97 ± 0.21 |
| Thin tubes (n = 50) | 64.6 | 51.7 ± 2.7 | 5.14 ± 0.17 |
| Wide tubes (n = 32) | 66.8 | 45.9 ± 3.6 | 5.00 ± 0.22 |
P > 0.05 for all comparisons by two-sided t tests.
See Fig. 1 c-e for examples.
Standard error of the mean.
As a further test, we compared CSX and CSX6 tags in the same glands of four uteri heterozygous for a single-nucleotide polymorphism (G or T) in CSX6 (Fig. 2). Consistent with random replication errors, there was little correlation between percent methylation at the two different CSX regions within individual glands (Fig. 5a). However, uterine averages correlated, consistent with the assumption that total methylation errors reflect stem cell mitotic ages. Although CSX6 demonstrated a slower increase in methylation with aging (a lower error rate), CSX6 methylation patterns were consistent with the CSX model, even when analyzing CSX6 alleles separately (Fig. 5b). Similar numbers of each CSX6 allele (G or T) were sequenced per gland, indicating that tag sampling was not greatly biased (Fig. 5c).
Fig. 5.
CSX6 endometrial tags. (a) Comparisons of CSX vs. CSX6 tags sampled from the same glands (“X”) reveal little correlation (gray line, correlation coefficient of 0.27), consistent with independent errors between CpG sites. However, average CSX and CSX6 uterine values (circles) correlated (black line, correlation coefficient of 0.99), consistent with the hypothesis that different CpG sites overall record similar numbers of divisions. (b) CSX6 methylation in four uteri (Patients 1, 9, 10, and 30) heterozygous for a single-nucleotide polymorphism. Circles represent averages of 12 tags, and triangles represent averages of six T (red) or G (blue) alleles. More stochastic scatter is expected with the smaller number of sampled T or G alleles. Simulations of nulliparous women for either two alleles per cell (gray lines) or one allele per cell (dotted lines), adjusted for different numbers of sampled CSX6 alleles and a lower methylation error rate (2 × 10-5 per CpG site per division), but with the same numbers of niche stem cells, mitotic rates, and niche turnover as the CSX simulations (Fig. 4), were consistent with the CSX6 experimental data. (c) Ratios of numbers of T to T+G alleles sampled from each gland. Skewed sampling bias (all one or the other allele) was not evident as approximately equal numbers of each allele were sampled.
Discussion
Human epithelial stem cell studies are limited because of the lack of specific markers or ability to distinguish between multiple stem cells that coexist within a gland. Here we use a molecular clock approach that infers numbers of divisions since birth and stem cell lineage survival from present-day endometrial gland methylation patterns. All somatic cells are related through a phylogenetic tree originating from the zygote, and adult stem cells may be defined as progenitors or common ancestors of differentiated cells (Fig. 1a). Most CpG islands are unmethylated at birth (11), and methylation potentially records divisions that occur during somatic cell tree expansion. Although stem cells cannot be directly isolated, and most methylation is sampled from more numerous differentiated cells, the patterns primarily reflect stem cell errors, because new errors in differentiated cells cannot accumulate. Endometrial glands are clonal (12), and differentiated cells may be separated from each other and their stem cells by relatively few divisions. Consistent with such a stem cell hierarchy, significant methylation differences were not found between gland fragments of different shapes (Table 3).
The mechanisms underlying methylation with aging are uncertain but likely stochastic, because errors are different between glands within a single uterus (Fig. 2). Although stochastic errors are difficult to interpret, endometrium sampled from different women can test whether average numbers of errors are proportional to total numbers of divisions since birth, because menstruation changes with age. Consistent with a somatic epigenetic clock, methylation paralleled mitotic age with a rapid increase before and relative stability after menopause (Fig. 3).
Estrogen appears to act as a carcinogen by stimulating cell proliferation in estrogen-responsive tissues (1). Progesterone may also play a role by opposing estrogen (1, 13). Parity decreases the risk of cancer by reducing numbers of menstrual cycles, and obesity increases the risk of cancer through elevated endogenous estrogen levels (1, 13, 14). Consistent with this biology, average methylation (and by inference stem cell divisions) was significantly less in multiparous and lean women >52 years of age. Methylation was 19% less in older multiparous women and increased 31% by obesity (Table 2). Mitotic age differences are expected to be small, because menstruation stops for only ≈1 year with each birth, and obesity does not appear to increase division beyond rates normally observed during the follicular phase of the menstrual cycle (13).
Relative numbers of divisions may be inferred through an obstetrical history, but methylation potentially records both mitotic age and ancestry, because epigenetic patterns are copied and physically passed from generation to generation. Endometrial glands are small and clonal (12) but could become polymorphic if maintained by multiple long-lived stem cell lineages. Individual glands were diverse populations with about five unique tags among eight sampled tags. However, rather than immortal stem cell lineages that always divide asymmetrically to produce one stem cell and one differentiated daughter, niches in which stem cell numbers rather than lineages remain constant (Fig. 4) are thought to maintain mammalian tissues (2, 15). Modeling indicated that the endometrial methylation patterns were also more consistent with niches containing multiple stem cells (see Supporting Text for details). The exact size of the endometrial niche cannot be determined from the data, but stem cells appear to be a minority of all cells. Fewer than 1% of endometrial cells exhibit clonogenic stem cell behavior in vitro (16). If a niche contains 64 stem cells, asymmetric stem cell division occurs ≈98% of the time. Other niche sizes and rates of asymmetric division are also compatible with the data (Fig. 4). Even though random niche stem cell loss with replacement eventually leads to the loss of all stem cell lineages except one, endometrial stem cell lineages are relatively long-lived, because this turnover is estimated to recur after a median interval of ≈10,000 divisions. Therefore, stem cell lineages persist through many menstrual cycles, because gland “bottlenecks” would recur about every 27 years, assuming on average one division per day after menarche. By comparison, human colon crypt niche stem cell lineages appear to be shorter-lived and turn over approximately every 8 years (9). There was no evidence for a decrease in niche stem cell number or survival with aging, because diversity was stable after menopause (Fig. 3).
Sequences have become useful tools to study evolution, because ancestry can be traced despite diverse morphologies and geographic locations (7). Theoretically the same benefits can be translated to compare different human tissues. Colon crypts exhibit less CSX tag diversity and a slower increase in age-related methylation (9) compared with endometrial glands (Fig. 6). Although further experiments are required to fully understand how methylation changes with division, a priori, by assuming similar error rates in different tissues, stem cells divide less frequently and have greater probabilities of extinction in the colon compared with endometrium. A greater ability of endometrial niche stem cells to persist may help explain the propensity of endometrium to survive outside the uterus, leading to endometriosis. Greater stem cell division rates may help explain why endometrial cancer is common [about half the incidence of colorectal cancer (17)] despite the small size of the uterus (<10 cm) relative to the much larger (>1-m) colon.
Fig. 6.
Endometrial gland vs. colon crypt CSX methylation. Colon crypt methylation (green circles, data from ref. 9) was linear with age and increased at a slower rate compared with endometrium, consistent with a relatively slower colon stem cell division rate. Tag diversity (unique tags per eight sampled tags) was lower in colon crypts compared with endometrial glands, consistent with a greater ability of endometrial stem cells to persist within a niche. Colon and endometrial diversity was stable with aging, suggesting niche stem cell numbers and turnover do not change with age.
A significant association between percent methylation and clinical parameters thought to influence endometrial divisions is empirical evidence that somatic cell histories are written within individual genomes. It is unlikely that our model fully translates the information encoded by random replication errors, and further studies are necessary to better define endometrial stem cell mitotic ages and ancestry. Species are typically analyzed at multiple different loci, and comparable comparisons between CSX and CSX6 methylation help support a clock-like link between age-related methylation and mitotic age and niche rather than immortal stem cell lineages. Many other factors, such as exogenous estrogen therapy (hormone replacement, oral contraceptives) and polymorphisms in hormonal signaling pathways, influence endometrial proliferation (1) but were not examined in this study. Variations in methylation among women (Fig. 3) may reflect these additional factors or scatter inherent in stochastic errors. Cell proliferation may be a major carcinogenic mechanism (14), but its relative importance and interactions with other risk factors are uncertain because of the inability to count total numbers of divisions directly. Endometrium facilitates testing of the idea that random replication errors accumulate in a clock-like manner, but conceptually all cell types surreptitiously record ancestry whenever they copy their genomes. The ability to infer mitotic ages from somatic genome drift may help unravel exactly how cell proliferation contributes to aging and diseases.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant DK61140 (to D.S.). S.T. is a Royal Society-Wolfson Research Merit Award holder and is supported in part by a grant from Cancer Research UK.
Author contributions: D.S. designed research; J.Y.K. and D.S. performed research; J.Y.K., S.T., and D.S. analyzed data; and S.T. and D.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: BMI, body mass index.
References
- 1.Pike, M. C., Pearce, C. L. & Wu, A. H. (2004) Oncogene 23, 6379-6391. [DOI] [PubMed] [Google Scholar]
- 2.Spradling, A., Drummond-Barbosa, D. & Kai, T. (2001) Nature 414, 98-104. [DOI] [PubMed] [Google Scholar]
- 3.Padykula, H. A. (1991) Ann. N.Y. Acad. Sci. 622, 47-56. [DOI] [PubMed] [Google Scholar]
- 4.Gargett, C. E. (2004) Aust. N. Z. J. Obstet. Gynaecol. 44, 380-386. [DOI] [PubMed] [Google Scholar]
- 5.Kondo, M., Wagers, A. J., Manz, M. G., Prohaska, S. S., Scherer, D. C., Beilhack, G. F., Shizuru, J. A. & Weissman, I. L. (2003) Annu. Rev. Immunol. 21, 759-806. [DOI] [PubMed] [Google Scholar]
- 6.Potten, C. S. & Loeffler, M. (1990) Development (Cambridge, U.K.) 110, 1001-1020. [DOI] [PubMed] [Google Scholar]
- 7.Bromham, L. & Penny, D. (2003) Nat. Rev. Genet. 4, 216-224. [DOI] [PubMed] [Google Scholar]
- 8.Ahuja, N., Li, Q., Mohan, A. L., Baylin, S. B. & Issa, J. P. (1998) Cancer Res. 58, 5489-5494. [PubMed] [Google Scholar]
- 9.Yatabe, Y., Tavaré, S. & Shibata D. (2001) Proc. Natl. Acad. Sci. USA 98, 10839-10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potten, C. S., Kellett, M., Rew, D. A. & Roberts, S. A. (1992) Gut 33, 524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird, A. (2002) Genes Dev. 16, 6-21. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka, M., Kyo, S., Kanaya, T., Yatabe, N., Nakamura, M., Maida, Y., Okabe, M. & Inoue, M. (2003) Am. J. Pathol. 163, 295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Key, T. J. & Pike, M. C. (1988) Br. J. Cancer 57, 205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preston-Martin, S., Pike, M. C., Ross, R. K., Jones, P. A. & Henderson, B. E. (1990) Cancer Res. 50, 7415-7421. [PubMed] [Google Scholar]
- 15.Watt, F. M. & Hogan, B. L. (2000) Science 287, 1427-1430. [DOI] [PubMed] [Google Scholar]
- 14.Chan, R. W., Schwab, K. E. & Gargett, C. E. (2004) Biol. Reprod. 70, 1738-1750. [DOI] [PubMed] [Google Scholar]
- 16.Jemal, A., Murray, T., Ward, E., Samuels, A., Tiwari, R. C., Ghafoor, A., Feuer, E. J. & Thun, M. J. (2005) CA Cancer J. Clin. 55, 10-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.