Abstract
The aryl hydrocarbon receptor (AHR) plays a role in three areas of biology that include the adaptive metabolism of xenobiotics, the toxic responses associated with exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (dioxin), and vascular remodeling of the developing embryo. To test the hypothesis that receptor signaling in different cell types is responsible for these aspects of AHR biology, we generated a conditional Ahr allele where exon 2 is flanked by loxP sites. Through the use of Cre-lox technology, we then investigated the role of AHR signaling in hepatocytes or endothelial cells in mediating prototypical endpoints of adaptive, toxic, or developmental signaling. Using this model, we provide evidence that AHR signaling in endothelial/hematopoietic cells is necessary for developmental closure of the ductus venosus, whereas AHR signaling in hepatocytes is necessary to generate adaptive and toxic responses of the liver in response to dioxin exposure. Taken together, these data illustrate the importance of cell-specific receptor signaling for the generation of distinct AHR-dependent physiological outcomes.
Keywords: Cre recombinase, ductus venosus, endothelial cell, hepatocyte, dioxin
The aryl hydrocarbon receptor (AHR) is a basic helix-loop-helix/Per-Arnt-Sim protein that plays an essential role in three areas of biology. In response to polycyclic aromatic hydrocarbons, the AHR up-regulates a battery of xenobiotic metabolizing enzymes that include the cytochromes P450, CYP1A1, CYP1A2, and CYP1B1 as well as the phase II enzymes GST-A1 and UGT1-06 (1, 2). In response to halogenated-dibenzo-p-dioxins, AHR activation results in the induction of xenobiotic metabolism plus a variety of toxic responses that include hepatocellular damage, thymic involution, epithelial hyperplasia, teratogenesis, and cancer (3-6). Finally, in response to an unknown developmental cue, the AHR influences normal vascular development, most notably the closure of a fetal vascular structure known as the ductus venosus (DV) (3, 7-9).
The mouse liver is a powerful model for investigations related to AHR biology. The mouse system allows the production of recombinant loci by gene targeting, whereas the liver provides a representation of each of the three aspects of AHR signal transduction. Using this model, we have provided evidence to suggest that the intracellular details of AHR signal transduction are similar for the adaptive, toxic, and developmental pathways. Through the use of recombinant Ahr and Arnt alleles, we have shown that AHR activation, AHR translocation to the nucleus, AHR dimerization with the aryl hydrocarbon receptor nuclear translocator (ARNT), and AHR-ARNT binding to dioxin responsive elements within the genome are required for adaptive metabolism, dioxin toxicity, and closure of the DV within the developing liver (3, 4, 7, 8, 10).
The question of how the AHR is able to produce multiple biological events from a similar signal transduction mechanism remains unclear. We hypothesize that receptor signaling in distinct cell types is responsible for these various aspects of AHR biology. To test this idea, we generated a conditional Ahr allele and used this model to investigate the importance of different cell populations and tissue specificity with respect to the role of AHR in mediating these endpoints. Specifically, through the use of Cre-lox technology, we have asked what effect AHR activation in hepatocytes or in endothelial cells has on the developmental, adaptive, and toxic pathways of AHR signaling in the liver.
Materials and Methods
Generation of Conditional Ahrfx Mice. The conditional Ahr allele (Ahrfx) contains exon 2 of Ahr flanked by loxP sites (”floxed”) for later excision by a cell-specific Cre recombinase (Cre). The Ahrfx mice were generated from the Ahrfxneo allele (8). Mice homozygous for the Ahrfxneo allele were crossed to a transgenic line carrying Cre under the control of the Ella promoter (CreElla, strain name: FVB/N-Tg(EIIa-cre)C5379Lmgd/J) (The Jackson Laboratory) to generate partial recombinants and obtain the conditional Ahrfx allele lacking the neomycin gene (11, 12). Discrimination between the WT and floxed allele was accomplished by PCR amplification of the region surrounding the loxP site located 3′ to exon 2, as described (8). Excision of the neomycin gene was detected by Southern blot analysis of BglII-digested genomic DNA by using a 500-bp probe specific to exon 2 of Ahr. This probe was amplified from PL1737 by using the primer set OL4676/4677 (8). Expression of the AHR protein was evaluated in mice homozygous for either the Ahr+ allele (Ahr+/+) or the conditional Ahrfx allele (Ahrfx/fx) by Western blot analysis of liver cytosolic fractions (4, 13).
After confirmation of neomycin excision, mice carrying the floxed exon 2 and the Cre transgene (Ahrfx/+CreElla) were then backcrossed to C57BL/6J to remove the CreElla transgene and produce the parental line (Ahrfx/+). Because of the fact that the conditional Ahrfx allele was generated originally from 129SvJ ES cells that carry the lower affinity Ahrd allele, we used a C57BL/6J strain congenic for DBA2-derived Ahrd allele to perform all backcrosses (14). The Ahrfx mice were backcrossed for at least four generations before experimental use.
Cell-Specific Excision of Ahrfx. To obtain mice harboring hepatocytes or endothelial cells with excision at Ahr, mice expressing the Ahrfx allele were crossed to mice expressing a Cre transgene driven by either the albumin promoter (CreAlb, strain name: B6.Cg-Tg(Alb-cre)21Mgn/J) or the Tie2 kinase promoter/enhancer (Tek) (CreTek, strain name: B6.Cg-Tg(Tek-cre)12Flv/J) (The Jackson Laboratory) (15, 16). Mice carrying either the CreAlb or CreTek transgene were backcrossed to C57BL/6J mice for seven generations before crossing with the conditional Ahrfx/fx mice. Mice homozygous for the floxed allele and hemizygous for the Cre transgene (Ahrfx/fxCreAlb or Ahrfx/fxCreTek) were obtained by crossing Ahrfx/+CreAlb or Ahrfx/+CreTek mice to Ahrfx/fx mice. Littermates that were negative for the Cre transgenes (Ahrfx/fx) were used as experimental controls. Because CreTek activity results in the deletion of floxed targets in the female germ line, male mice expressing the conditional Ahrfx allele and the CreTek transgene were used to transmit CreTek to the offspring (15).
Genotyping for the Cre transgene was performed by PCR on DNA isolated from tail biopsies by using the forward primer, OL2642 (5′-TGCCTGCATTACCGGTCGATGC) and reverse primer, OL2643 (5′-CCATGAGTGAACGAACCTGGTCG) in a reaction consisting of 2.5 units Taq polymerase (Promega), 50 mM KCl, 10 mM Tris·HCl (pH 9 at 25°C), 1.5 mM MgCl2, 1% Triton X-100, 200 μM dNTPs, and 0.1 μM each primer. The PCR was carried out for 30 cycles (95°C/30 s; 60°C/30 s; 72°C/30 s). A 450-bp band confirmed the presence of the Cre transgene. Samples negative for Cre did not amplify a product.
Analysis of Ahrfx excision was carried out by multiplex PCR by using the forward primers OL4062 (5′-GTCACTCAGCATTACACTTTCTA) and OL4064 (5′-CAGTGGGAATAAGGCAAGAGTGA) in combination with the reverse primer OL4088 (5′-GGTACAAGTGCACATGCCTGC). PCR conditions are similar to those described (8). The Ahrfx-excised allele (OL4062/4088) amplified a 180-bp band, whereas amplification from the Ahrfx-unexcised allele (OL4064/4088) resulted in a 140-bp band. The WT allele, if present, generated a 106-bp band (OL4064/OL4088).
To evaluate the specificity of excision events, hepatocytes were separated from nonparenchymal cells by digestion of the liver with Liberase Blendzyme (Roche) followed by centrifugation over a two-step Percoll gradient (17). Genomic DNA from various cell types was isolated by using the DNeasy tissue kit (Qiagen, Valencia, CA).
Toxicology Studies. Mice were housed in a selective pathogen-free facility on corncob bedding with food and water ad libitum according to the rules and guidelines set by the University of Wisconsin. In toxicology studies, 5-wk-old male mice were dosed by i.p. injection once per week for 4 wk with 100 μg/kg dioxin in DMSO or with DMSO alone. This dose of dioxin was necessary to elicit toxic endpoints in the conditional Ahrfx/fx mice, which carry the low affinity form of the receptor (14). After (7 d) the final injection of dioxin, animals were anesthetized by inhalation with ethyl-ether for blood draw and then killed by cervical dislocation for organ harvest. Whole blood was obtained by retro-orbital puncture for analysis of serum alanine aminotransferase (ALT), which was performed by the Clinical Pathology Laboratory (University of Wisconsin, School of Veterinary Medicine). Liver and thymus were dissected and weighed. Sections from the left lobe of the liver were fixed in 10% formalin and embedded in paraffin for staining with hematoxylin and eosin. The remaining liver tissue was immersed in RNAlater solution (Qiagen) and then stored at -80°C until total RNA was prepared as described in Expression Analysis.
Expression Analysis. Total liver RNA was prepared by using the RNeasy Protect system (Qiagen). The quality and quantity of RNA was determined on an Agilent 2100 Bioanalyzer by using the RNA Nano Labchip (Palo Alto, CA). The RNA samples were typically at a concentration of >1 μg/ml and the 260/280 absorbance ratio was between 1.7 and 2. For expression analysis, Northern blot hybridization was performed on 10 μg of total liver RNA by using cDNA probes specific to mouse Cyp1a1 (PL2051), Cyp1a2 (PL1232), Cyp1b1 (PL2129), and rat Gapdh (PL2031).
Assessment of DV Status. The status of the DV in Ahrfx/fx, Ahrfx/fxCreAlb, and Ahrfx/fxCreTek mice was assessed by perfusion of the liver with trypan blue and confirmed by time-lapsed angiography as described (8).
Statistical Analysis. In situations where multiple comparisons could be made, an ANOVA followed by Tukey's test was performed. For genotype frequencies, Fisher's exact test was used.
Results
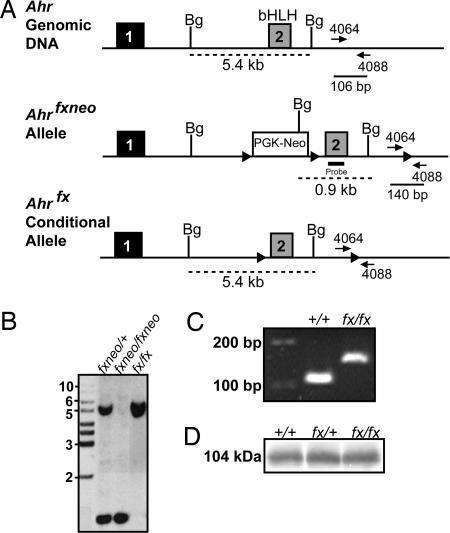
Generation of Conditional Ahrfx Mice. Mice harboring the conditional Ahrfx allele were generated from the Ahrfxneo allele by excision of the neomycin gene through crosses with mice expressing the CreElla transgene (8, 11, 12). A map of both the Ahrfxneo and conditional Ahrfx alleles is shown in Fig. 1A. For comparison, a partial map of the Ahr structural gene is also shown. Generation of the Ahrfx allele lacking neomycin was confirmed by both Southern blot and PCR analysis of genomic DNA (Fig. 1 B and C).
Fig. 1.
Generation of the conditional Ahrfx mice. (A) Schematic diagram illustrating the region surrounding the basic helix-loop-helix (bHLH) domain of the murine Ahr locus, the Ahrfxneo (fxneo) allele, and the conditional Ahrfx (fx) allele. Exon numbers reflect known coding exons. To generate the conditional Ahrfx allele, the CreElla mouse was crossed to the Ahrfxneo mouse. Selective excision of the neomycin gene was detected by using a combination of Southern blotting (B) and PCR genotyping (C) of genomic DNA. Dashed lines indicate fragment sizes detected by the Southern probe after digestion of 10 μg of genomic tail DNA with BglII. Solid lines represent the fragment sizes generated by PCR amplification of the WT (+/+) and Ahrfx/fx (fx/fx) alleles by using OL4064 as the forward primer and OL4088 as reverse primer. (B) Southern blot of mouse tail biopsies showing a 0.9-kb band indicating the presence of the neomycin gene in the Ahrfxneo allele and 5.4-kb band detecting the WT or Ahrfx/fx allele. (C) PCR genotyping of tail biopsies showing bands of 106 bp and 140 bp generated from the amplification of the WT and Ahrfx/fx alleles, respectively. (D) Western blot showing AHR expression in liver from WT, heterozygous Ahrfx/+, and conditional Ahrfx/fx mice.
As a result of the deletion of the neomycin gene, protein expression from the conditional Ahrfx/fx allele was found to be equivalent to WT levels by Western blot analysis (Fig. 1D). Liver perfusion studies on homozygous Ahrfx/fx mice demonstrated that 100% of mice (19/19) displayed normal DV closure and normal liver perfusion, similar to that of WT mice (data not shown).
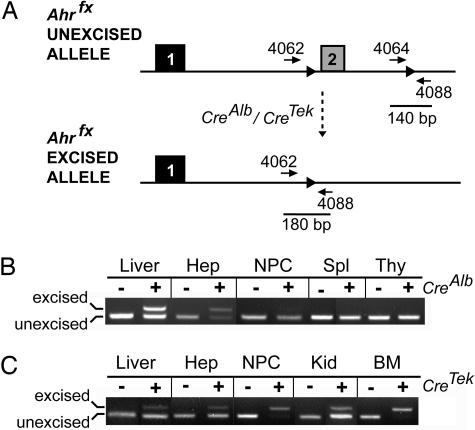
Cell-Specific Excision of the Conditional Ahrfx Allele. To investigate the importance of cell-specific AHR signaling, we asked whether the AHR in hepatocytes or endothelial cells was responsible for the prototypical endpoints of adaptive, toxic, or developmental AHR biology. To this end, we began by generating mice in which the Ahr was deleted in hepatocytes or endothelial cells (Fig. 2A).
Fig. 2.
Specificity of CreAlb- and CreTek-mediated excision of the Ahrfx allele. (A) Schematic illustration of the Ahrfx-unexcised and the Ahrfx-excised alleles. Solid lines represent the fragment sizes generated by PCR amplification of the Ahrfx-unexcised and Ahrfx-excised alleles by using the forward primers OL4062 and OL4064 and the reverse primer OL4088. (B) Specificity of Ahrfx excision by CreAlb was determined by genotyping for both the unexcised and excised alleles of Ahrfx in genomic DNA from various tissues obtained from Ahrfx/fx and Ahrfx/fxCreAlb mice. (C) Specificity of Ahrfx excision by CreTek was determined by genotyping for both the unexcised and excised alleles of Ahrfx in genomic DNA from various tissues obtained from Ahrfx/fx and Ahrfx/fxCreTek mice. Liver, whole liver; Hep, hepatocytes; NPC, hepatic nonparenchymal cells; Spl, spleen; Kid, kidney; Thy, thymus; BM, bone marrow.
To examine the specificity of excision events in Ahrfx/fxCreAlb mice, we analyzed various tissues for the presence of both the Ahrfx-unexcised and Ahrfx-excised alleles. In the absence of CreAlb, the Ahrfx/fx mice showed only the Ahrfx-unexcised allele in every tissue examined (Fig. 2B and data not shown). In the presence of CreAlb, the Ahrfx-excised allele was observed only in the liver (Fig. 2B and data not shown). The Ahrfx-unexcised allele was also observed in the liver, suggesting that Ahr excision did not occur in the nonparenchymal cells of this organ. To investigate this further, hepatocytes and nonparenchymal cells were separated from Ahrfx/fx and Ahrfx/fxCreAlb mouse livers, and excision events were examined in both fractions. In keeping with what is known about CreAlb expression in hepatocytes, the major band amplified in the nonparenchymal cell preparations represented the Ahrfx-unexcised allele (Fig. 2B). In hepatocytes, the predominant band observed represented the Ahrfx-excised allele. The presence of the Ahrfx-unexcised allele in the parenchymal fraction may indicate contamination by nonparenchymal cells or that a small fraction of hepatocytes are still expressing AHR in this system (Fig. 2B).
To examine the specificity of excision events in Ahrfx/fxCreTek mice, we again analyzed various tissues for the presence of both the Ahrfx-unexcised and Ahrfx-excised alleles (Fig. 2C). Because of the presence of endothelial cells in almost all tissues and the fact that CreTek is also expressed in cells of hematopoietic lineage, both the Ahrfx-unexcised and Ahrfx-excised alleles were detected in all organs examined, including liver, spleen, kidney, heart, lung, thymus, and bone marrow (Fig. 2C) (15). For this reason, we assessed the relative excision in hepatic parenchymal and nonparenchymal cell fractions under nonsaturating PCR conditions. Under these conditions, the major band amplified in the nonparenchymal cell fraction of the liver represented the Ahrfx-excised allele (Fig. 2C). In contrast, the major band amplified from the hepatocyte fraction represented the Ahrfx-unexcised allele (Fig. 2C). In the absence of CreTek, the Ahrfx/fx mice showed only the Ahrfx-unexcised allele in all tissues examined (Fig. 2C).
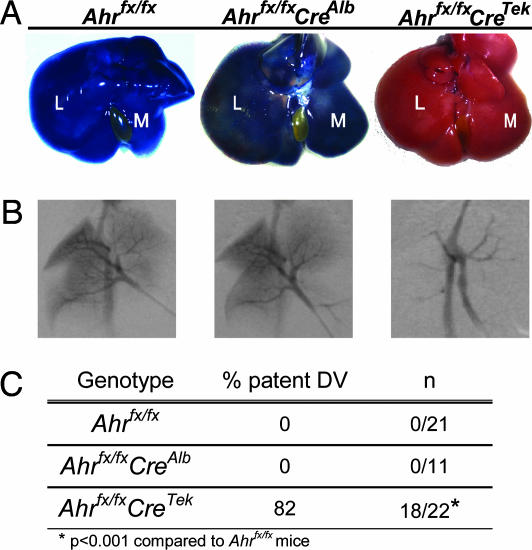
Developmental Failure of DV Closure in Ahrfx/fxCreTek Mice. To assess the importance of cell-specific AHR expression on developmental aspects of signaling, we used our conditional allele in experiments examining a prototypical endpoint, DV patency. Because of the persistence of a patent DV in the liver of various Ahr mutants, we asked what role hepatocytes and endothelial cells played in normal developmental closure of this structure (3, 7-9). To this end, we generated two cell-specific excisions of AHR by crossing the conditional Ahrfx/fx mice to either CreAlb or CreTek mice. We then compared the effect of these genotypes on DV status in adult mice by assessment of liver vasculature by using trypan blue perfusion and angiography (Fig. 3 A and B). In a manner similar to WT mice described previously, none (0/21) of the unexcised Ahrfx/fx mice retained a patent DV as adults (Fig. 3 A-C) (8). Similarly, depletion of hepatocyte AHR did not alter the frequency of DV patency compared with the Ahrfx/fx mice, with none (0/11) of the Ahrfx/fxCreAlb mice demonstrating a patent DV (Fig. 3 A-C). In contrast to the Ahrfx/fx mice, ≈80% (18/22) of the Ahrfx/fxCreTek mice displayed a patent DV (P < 0.001, Fig. 3 A-C). Liver angiography confirmed that the DV remains open in the majority of Ahrfx/fxCreTek mice, whereas the Ahrfx/fx and Ahr fx/fxCreAlb mice show normal liver perfusion and DV closure (Fig. 3B).
Fig. 3.
The DV fails to close in Ahrfx/fxCreTek mice. (A) Representative examples of livers from Ahrfx/fx, Ahrfx/fxCreAlb, and Ahrfx/fxCreTek mice perfused with trypan blue. Livers were cannulated via the portal vein, flushed with PBS, and then injected with 0.5 ml of trypan blue. Upon perfusion of a normal liver with trypan blue, complete filling of the hepatic vasculature resulted in a blue coloration and indicated normal DV closure. In contrast, failure of the liver to turn blue indicated porto-systemic shunting and a patent DV. M, median lobe; L, left lobe. Representative angiographs of livers (B) and frequency of DV patency (C) in adult Ahrfx/fx, Ahrfx/fxCreAlb, and Ahrfx/fxCreTek mice.
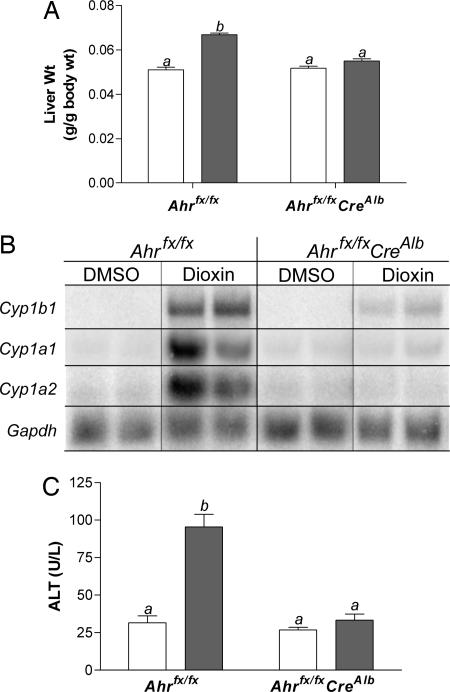
Hepatocytes Are Major Contributors to the Adaptive Response in the Liver. To investigate the role of the hepatocyte in generating the liver's adaptive metabolic response, we began by examining the influence of dioxin on transcriptional up-regulation in livers from mice with and without hepatocyte AHR. After dioxin treatment, control and treated livers were weighed, and total RNA was used for Northern blot analysis. In Ahrfx/fx mice, liver weights increased 30% by dioxin treatment compared with vehicle-treated controls (P < 0.05, Fig. 4A). In contrast, dioxin exposure failed to induce hepatomegaly in Ahrfx/fxCreAlb mice. Northern blot analysis confirmed that the dioxin-regulatable genes, Cyp1a1, Cyp1a2, and Cyp1b1 are all markedly induced in Ahrfx/fx mice treated with dioxin compared with vehicle-treated Ahrfx/fx mice (Fig. 4B). After dioxin treatment, Cyp1a1 expression in livers of Ahrfx/fxCreAlb mice is weakly induced compared with that observed in livers of Ahrfx/fx mice, indicating that AHR activation in hepatocytes is the major contributor to Cyp1a1 induction in response to dioxin. In DMSO-treated mice, basal expression of Cyp1a2 is independent of the presence of hepatocyte AHR. In mice lacking hepatocyte AHR (Ahrfx/fxCreAlb mice), induction of Cyp1a2 expression in the liver after dioxin treatment is abolished; however the low level basal Cyp1a2 band remains. Finally, after dioxin treatment, partial induction of Cyp1b1 expression is observed in livers of Ahrfx/fxCreAlb mice compared with that found in Ahrfx/fx mice, suggesting that AHR in nonparenchymal cells is a significant contributor to the total Cyp1b1 induction in this organ.
Fig. 4.
Adaptive metabolism and hepatocellular toxicity are dependent on AHR signaling in hepatocytes. Mice were treated once per week for 4 wk with 100 μg of dioxin/kg in DMSO or DMSO alone and then killed 7 d after the final injection. White bars, vehicle-treated animals; gray bars, dioxin-treated animals. The Ahrfx/fxCreAlb groups each contain eight animals, whereas the vehicle-treated Ahrfx/fx group contains four animals, and the dioxin-treated Ahrfx/fx group contains 12 animals. Error bars, SE. Those groups not sharing a superscript letter differ significantly at P ≤ 0.05. (A) Liver weights in Ahrfx/fx and Ahrfx/fxCreAlb mice in response to dioxin treatment. (B) Representative Northern blot analysis of Cyp1a1, Cyp1a2, and Cyp1b1 expression in Ahrfx/fx and Ahrfx/fxCreAlb mice in response to dioxin treatment. Gapdh expression was used as a loading control. (C) ALT values in Ahrfx/fx and Ahrfx/fxCreAlb in response to dioxin treatment.
Liver Toxicity Is Hepatocyte-Dependent. As further support of the cell specificity of dioxin-induced liver toxicity, we examined several pathological endpoints in mice lacking hepatocyte AHR. As one method to assess hepatocellular damage, we measured serum ALT in animals exposed to dioxin. After dioxin treatment, the Ahrfx/fx mice displayed ALT values that were 3-fold higher than the Ahrfx/fxCreAlb mice (P < 0.05, Fig. 4C). In contrast, serum ALT levels in mice lacking hepatocyte AHR that were exposed to dioxin were not significantly different from DMSO-treated controls.
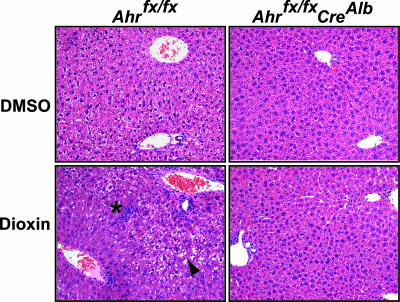
As an independent measure of hepatotoxicity, liver sections from dioxin-treated and control mice were examined for histological evidence of inflammation and hydropic changes, both classic endpoints of dioxin exposure (18). In comparison to their vehicle treated controls, analysis of the hematoxylin and eosin stained liver sections revealed that the conditional Ahrfx/fx mice treated with dioxin displayed extensive hydropic vacuolation of the portal region characterized by the presence of clear cytoplasmic elements with a ragged appearance (Fig. 5). In addition, these mice also displayed areas of mild congestion and extensive pyogranuloma formation, indicating inflammation and neutrophil infiltration (Fig. 5). In contrast, liver sections from Ahrfx/fxCreAlb mice treated with dioxin displayed decreased zonal vacuolation of the parenchyma and fewer pyogranulomas in comparison to the dioxin-treated Ahrfx/fx mice (Fig. 5).
Fig. 5.
Dioxin-induced hepatocellular toxicity depends on AHR signaling in hepatocytes. Hepatocellular toxicity in dioxin-treated Ahrfx/fxCreAlb mice is attenuated compared with dioxin-treated Ahrfx/fx mice. Liver sections were stained with hematoxylin and eosin for assessment of liver pathology after dioxin treatment regimen. Black arrowhead, extensive hydropic vacuolation and degeneration in portal region; asterisk, granuloctye infiltration. (Magnification: ×20.)
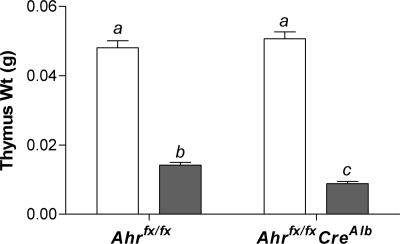
Dioxin-Induced Thymic Involution Is Independent of Hepatocyte AHR Signaling. To demonstrate that extrahepatic toxicity can be independent of hepatocellular AHR, we also quantified thymic involution, a well established marker of dioxin exposure (19, 20). Thymus weights were measured after dioxin treatment (Fig. 6). The Ahrfx/fx mice, with unexcised hepatocyte AHR, showed a 72% reduction in thymus weight as a result of dioxin exposure compared with their vehicle-treated controls (P < 0.05). The dioxin-treated Ahrfx/fxCreAlb mice, with excised hepatocyte AHR, showed an 82% decrease in thymus weight compared with controls (P < 0.05). DMSO-treated Ahrfx/fxCreAlb mice showed no difference in thymus weight compared with Ahrfx/fx controls.
Fig. 6.
Dioxin-induced thymic involution is independent of AHR signaling in hepatocytes. Thymus weights from control and dioxin-treated Ahrfx/fx and Ahrfx/fxCreAlb were measured. White bars, vehicle-treated animals; gray bars, dioxin-treated animals. The Ahrfx/fxCreAlb groups each contain eight animals, whereas the vehicle-treated Ahrfx/fx group contains four animals, and the dioxin-treated Ahrfx/fx group contains 12 animals. Error bars, SE. Those groups not sharing a superscript letter differ significantly at P ≤ 0.05.
Discussion
The AHR controls the adaptive up-regulation of xenobiotic metabolizing enzymes in response to polycyclic aromatic hydrocarbons; it regulates the toxicity of halogenated dioxins, and it directs the developmental remodeling of vascular architecture in the liver. Recent experiments with Ahr and Arnt mutant mice suggest that the three distinct physiological outcomes of AHR signal transduction are generated from a similar series of intracellular steps that include receptor activation, translocation to the nucleus, dimerization with ARNT, and binding to dioxin responsive elements regulating target genes (3, 4, 8).
Given our idea that the major aspects of AHR signal transduction are the same for these three pathways, we set out to understand how signaling through one receptor could produce such different biological outcomes. To this end, we asked whether the cell specificity of AHR activation was an important determinant in the physiological outcome of AHR signal transduction. To examine this idea, we generated mice harboring a conditional allele of Ahr, designated Ahrfx. The conditional inactivation of the Ahrfx allele was then accomplished by Cre-mediated deletion of exon 2, which contains the region encoding the basic helix-loop-helix domain essential for DNA binding (Fig. 1 A) (21, 22). Deletion of this exon has been shown to completely eliminate receptor expression from this locus (10). Using the liver as a model system, we then asked how inactivation of AHR in various cell types affected the outcome of developmental, adaptive, and toxic signaling.
Developmental Closure of the DV Is Dependent on AHR Signaling in Endothelial/Hematopoietic Cells. As one measurable outcome of developmental AHR signaling, we assessed DV closure in mice harboring cell-specific excisions of the Ahr. The DV is a fetal structure designed to shunt blood from the umbilical vein directly to the inferior vena cava. In mice, vascular remodeling within 48 h of birth leads to DV closure thereby establishing the normal, adult, hepatic blood flow pattern (9). Mice expressing the null allele of Ahr, or the hypomorphic alleles of Ahr (Ahrfxneo) or Arnt (Arntfxneo), fail to undergo this hepatic vascular remodeling and display a patent DV as adults (3, 8, 9). To examine which cell types may be responsible for this biology, we examined the frequency of DV closure in Ahrfx/fxCreAlb and Ahrfx/fxCreTek mice. To this end, we demonstrated that ≈80% of adult Ahrfx/fx CreTek mice have a patent DV, a similar frequency to that seen in Ahr and Arnt mutants (Fig. 3) (3, 8, 9). In contrast, none of the adult Ahrfx/fxCreAlb mice have a patent DV, and they display liver perfusion similar to that seen in WT mice and the Ahrfx-unexcised mice (Fig. 3) (3, 8).
The vascular nature of the Ahr null phenotype and the reproduction of this phenotype in Ahrfx/fxCreTek mice lead us to speculate that it is receptor in endothelial cells that is responsible for DV closure. Yet, it is important to note that there are other cellular candidates that are not ruled out by this model system. Although not widely recognized, Tek is also known to drive Cre expression in cells of hematopoietic lineage (15). In support of this fact, extensive excision of the Ahrfx allele was observed in the bone marrow, spleen, and thymus obtained from Ahrfx/fxCreTek mice (Fig. 2B). Therefore, the presence of a patent DV in Ahrfx/fxCreTek mice, but not in Ahrfx/fxCreAlb mice, supports the idea of cell autonomy with respect developmental signaling of the AHR. However, future experiments are necessary to delineate the relative importance of endothelial cells vs. hematopoietic cells in this developmental pathway.
Based on these findings, we suggest a model to explain how AHR activation influences DV patency. Given this vascular phenotype, we make the simplifying assumption that receptor signaling in endothelial cells, and not hematopoietic cells, is essential for vascular remodeling and DV closure. In our model, endothelial cell AHR is important for sensing humoral or intracellular signals that regulate vascular tone during the developmental transition from fetal to adult hepatic vasculature. In turn, the activated receptor may up-regulate CYP1A monoxygenases that either clear or generate vasoactive compounds. The role of AHR could lie at the level of endothelial cells in hepatic sinusoids or directly on endothelial cells in the DV. Arguing for a sinusoidal event is the observation that adult mice lacking Ahr retain a fetal pattern of anastomotic hepatic sinusoids (7). Failure of the anastomotic sinusoids to resolve during development may increase hepatic resistance and portal hypertension preventing normal closure of the DV.
Hepatocellular AHR Signaling Is Essential for the Adaptive Capacity of the Liver. To investigate the relationship between cell specificity and the adaptive metabolic response, we evaluated expression levels from select members of the dioxin-responsive xenobiotic metabolizing enzyme gene battery. Mice lacking hepatocyte AHR (Ahrfx/fxCreAlb mice) failed to generate the marked induction of Cyp1a1, Cyp1a2, and Cyp1b1 in response to dioxin exposure that was observed in Ahrfx-unexcised mice. The weak level of Cyp1a1 and Cyp1b1 induction observed in Ahrfx/fxCreAlb mice treated with dioxin likely reflects the contribution coming from AHR activation in nonparenchymal cells of the liver (23). However, considering the large proportion of hepatocytes in whole liver, the contribution of hepatocellular AHR signaling to functional metabolic capacity is likely to be more significant. Given the normal closure of the DV, this animal model holds great promise as a model of first pass metabolism, especially with respect to the importance of dioxin responsive element-driven genes in xenobiotic disposition and pharmacology.
Dioxin-Induced Liver Toxicity Is Dependent on AHR Signaling in Hepatocytes. To identify the hepatic cell type with a major role in toxic signaling, we performed experiments in mice harboring the hepatocyte-specific excision of the AHR. The failure of dioxin-treated Ahrfx/fxCreAlb mice to display hepatomegaly, increases in serum ALT or significant pathological changes in the liver, clearly demonstrated that AHR signaling in hepatocytes is essential for the generation of toxic responses caused by dioxin exposure. The finding that dioxin-induced thymic involution is independent of hepatocyte signaling suggests that another unique cell type, presumably thymocytes, is responsible for dioxin-induced toxicity in that organ.
Based on these findings, we suggest a model of dioxin hepatotoxicity. In this model, AHR activation and subsequent target gene up-regulation occurs first in hepatocytes, which act as “primary responders”. The cellular effects of AHR activation in these primary responders leads to stimulation of secondary cell types that react to the hepatocyte stress. In such a model, the response to dioxin that began in hepatocytes is progressive, gradually recruiting nonparenchymal and possibly extra-hepatic cell types. As additional evidence for this model, we have previously described the dioxin response in mice that are compound nulls for the receptors for TNFα, TNFβ, IL1α, and IL1β (18). Using that model system, we were able to identify aspects of dioxin-induced hepatotoxicity that are dependent on IL1-like cytokines, a candidate secondary response to hepatocellular stress. In these compound null mice, dioxin treatment produces Cyp1a induction, hepatomegaly, and hydropic degeneration of the liver but fails to have an impact other measures of hepatocellular toxicity, namely serum ALT levels and infiltration of inflammatory cells. Thus, the toxic consequences of dioxin require AHR activation in the hepatocyte, but the full consequences of the toxic pathway of AHR signaling are only realized once the tissue or organism as a whole responds.
Conclusions. We have described the generation of a conditional Ahrfx allele and have used this model to investigate the importance of cell-specific receptor activation in three areas of AHR biology. Through the generation of Ahrfx/fxCreTek mice, we provide evidence to suggest that AHR activation and signaling in endothelial/hematopoietic cells is necessary for vascular remodeling and developmental closure of the DV. We then extend the idea of cell specificity of AHR biology by examining the response to dioxin treatment in mice lacking hepatocyte AHR. We found that Ahrfx/fxCreAlb mice treated with dioxin fail to generate a significant adaptive metabolic response or any of the classic endpoints of dioxin-induced hepatotoxicity. Therefore, the adaptive and toxic responses of the liver are dependent on AHR activation in hepatocytes and the developmental response depends on AHR activation in endothelial/hematopoietic cells. Taken in sum, the data provide evidence to support the idea that cell specificity of receptor activation is an important determinant of the physiological outcome of AHR signaling in the liver.
Acknowledgments
We thank Henry Pitot and Ruth Sullivan for their pathological expertise. This work was supported by National Institutes of Health Grants ES006883, CA014520, and CA022484. J.A.W. would like to acknowledge the postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada. This manuscript is dedicated to the memory of Jessie Alexander Rhoades Walisser, 1923-2005.
Author contributions: J.A.W. and C.A.B. designed research; J.A.W., E.G., K.P., and A.L.L. performed research; J.A.W. contributed new reagents/analytical tools; J.A.W. and C.A.B. analyzed data; and J.A.W. and C.A.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AHR, aryl hydrocarbon receptor; ALT, alanine aminotransferase; ARNT, aryl hydrocarbon receptor nuclear translocator; Cre, Cre recombinase; DV, ductus venosus; Tek, Tie2 kinase promoter/enhancer.
References
- 1.Schmidt, J. V. & Bradfield, C. A. (1996) Annu. Rev. Cell Dev. Biol. 12, 55-89. [DOI] [PubMed] [Google Scholar]
- 2.Hankinson, O. (1995) Annu. Rev. Pharmacol. Toxicol. 35, 307-340. [DOI] [PubMed] [Google Scholar]
- 3.Walisser, J. A., Bunger, M. K., Glover, E., Harstad, E. B. & Bradfield, C. A. (2004) J. Biol. Chem. 279, 16326-16331. [DOI] [PubMed] [Google Scholar]
- 4.Bunger, M. K., Moran, S. M., Glover, E., Thomae, T. L., Lahvis, G. P., Lin, B. C. & Bradfield, C. A. (2003) J. Biol. Chem. 278, 17767-17774. [DOI] [PubMed] [Google Scholar]
- 5.Tomita, S., Jiang, H., Ueno, T., Takagi, S., Tohi, K., Maekawa, S., Miyatake, A., Furukawa, A., Gonzalez, F. J., Takeda, J., et al. (2003) J. Immunol. 171, 4113-4120. [DOI] [PubMed] [Google Scholar]
- 6.Uno, S., Dalton, T. P., Sinclair, P. R., Gorman, N., Wang, B., Smith, A. G., Miller, M. L., Shertzer, H. G. & Nebert, D. W. (2004) Toxicol. Appl. Pharmacol. 196, 410-421. [DOI] [PubMed] [Google Scholar]
- 7.Lahvis, G. P., Lindell, S. L., Thomas, R. S., McCuskey, R. S., Murphy, C., Glover, E., Bentz, M., Southard, J. & Bradfield, C. A. (2000) Proc. Natl. Acad. Sci. USA 97, 10442-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walisser, J. A., Bunger, M. B., Glover, E. & Bradfield, C. A. (2004) Proc. Natl. Acad. Sci. USA 101, 16677-16682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahvis, G. P., Pyzaski, R. W., Glover, E., Pitot, H. C., McElwee, M. K. & Bradfield, C. A. (2004) Mol. Biol. Cell 67, 714-720. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, J. V., Su, G. H., Reddy, J. K., Simon, M. C. & Bradfield, C. A. (1996) Proc. Natl. Acad. Sci. USA 93, 6731-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., Alt, F. W. & Westphal, H. (1996) Proc. Natl. Acad. Sci. USA 93, 5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holzenberger, M., Lenzner, C., Leneuve, P., Zaoui, R., Hamard, G., Vaulont, S. & Bouc, Y. L. (2000) Nucleic Acids Res. 28, E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt, J. V., Carver, L. A. & Bradfield, C. A. (1993) J. Biol. Chem. 268, 22203-22209. [PubMed] [Google Scholar]
- 14.Poland, A. & Glover, E. (1980) Mol. Pharmacol. 17, 86-94. [PubMed] [Google Scholar]
- 15.Koni, P. A., Joshi, S. K., Temann, U. A., Olson, D., Burkly, L. & Flavell, R. A. (2001) J. Exp. Med. 193, 741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postic, C., Shiota, M., Niswender, K. D., Jetton, T. L., Chen, Y., Moates, J. M., Shelton, K. D., Lindner, J., Cherrington, A. D. & Magnuson, M. A. (1999) J. Biol. Chem. 274, 305-315. [DOI] [PubMed] [Google Scholar]
- 17.Braet, F., De Zanger, R., Sasaoki, T., Baekeland, M., Janssens, P., Smedsrod, B. & Wisse, E. (1994) Lab. Invest 70, 944-952. [PubMed] [Google Scholar]
- 18.Pande, K., Moran, S. M. & Bradfield, C. A. (2005) Mol. Pharmacol. 67, 1393-1398. [DOI] [PubMed] [Google Scholar]
- 19.Poland, A. & Knutson, J. C. (1982) Annu. Rev. Pharmacol. Toxicol. 22, 517-554. [DOI] [PubMed] [Google Scholar]
- 20.Holsapple, M. P., Morris, D. L., Wood, S. C. & Snyder, N. K. (1991) Annu. Rev. Pharmacol. Toxicol. 31, 73-100. [DOI] [PubMed] [Google Scholar]
- 21.Fukunaga, B. N., Probst, M. R., Reisz-Porszasz, S. & Hankinson, O. (1995) J. Biol. Chem. 270, 29270-29278. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga, B. N. & Hankinson, O. (1996) J. Biol. Chem. 271, 3743-3749. [DOI] [PubMed] [Google Scholar]
- 23.Piscaglia, F., Knittel, T., Kobold, D., Barnikol-Watanabe, S., Di Rocco, P. & Ramadori, G. (1999) Biochem. Pharmacol. 58, 157-165. [DOI] [PubMed] [Google Scholar]