Abstract
Defects of mitochondrial DNA (mtDNA) maintenance have recently been associated with inherited neurodegenerative and muscle diseases and the aging process. Twinkle is a nuclear-encoded mtDNA helicase, dominant mutations of which cause adult-onset progressive external ophthalmoplegia (PEO) with multiple mtDNA deletions. We have generated transgenic mice expressing mouse Twinkle with PEO patient mutations. Multiple mtDNA deletions accumulate in the tissues of these mice, resulting in progressive respiratory dysfunction and chronic late-onset mitochondrial disease starting at 1 year of age. The muscles of the mice faithfully replicate all of the key histological, genetic, and biochemical features of PEO patients. Furthermore, the mice have progressive deficiency of cytochrome c oxidase in distinct neuronal populations. These “deletor” mice do not, however, show premature aging, indicating that subtle accumulation of mtDNA deletions and progressive respiratory chain dysfunction are not sufficient to accelerate aging. This model is a valuable tool for therapy development and testing for adult-onset mitochondrial disorders.
Keywords: mouse model, progressive external ophthalmoplegia, mitochondrial DNA replication
Defects in nuclear-encoded proteins essential for mitochondrial DNA (mtDNA) maintenance have recently been shown to be important causes of mitochondrial disease. This disease group is characterized by progressive loss of mtDNA and/or somatic accumulation of multiple mtDNA deletions (1, 2). Defects of the mitochondrial replicative DNA polymerase γ (POLG) lead to a wide spectrum of neurological diseases, such as progressive external ophthalmoplegia (PEO), Alpers syndrome, parkinsonism and spinocerebellar ataxia (3–6). Furthermore, knock-in mice lacking the proofreading activity of POLG accumulated high amounts of mtDNA mutations and developed symptoms associated with advanced aging, supporting the role of mtDNA mutations also in normal aging (7).
We reported dominant mutations of the mitochondrial replicative helicase Twinkle in PEO with multiple mtDNA deletions (8). The disease manifested as myopathy, often affecting the extraocular, limb, and facial muscles, sometimes accompanied by major depression (9, 10). In vitro, Twinkle has been shown to form a minimal mtDNA replisome together with the mitochondrial single-stranded DNA-binding protein and POLG (11). We have recently demonstrated that Twinkle is essential for mtDNA maintenance because human cells devoid of Twinkle rapidly lost their mtDNA, and Twinkle has an active role in mtDNA copy number control in vivo (12). Twinkle bears homology to the bacteriophage T7 gp4 hexameric primase-helicase (8). PEO mutations are predicted to localize at or near the putative regions that link the subunits to the hexameric ring. Because the dNTP binding by the T7-type helicases occurs at the subunit interface, correct subunit interaction is essential for helicase activity (13, 14). Dominant mutations in Twinkle could therefore both enhance dNTP breakdown and affect the processivity of mtDNA replication (8).
No effective treatment for mtDNA disease is available, and development of therapeutic strategies has been hindered by the lack of disease models that allow intervention and follow-up. Mouse models with mitochondrial dysfunction have been generated (15–18), but their severe disease course resembled that of childhood diseases and no models for chronic late-onset disease have been available. We examined the consequences of dominant Twinkle-PEO mutations in transgenic mice. These mice carrying human patient mutations have gradual worsening of the respiratory chain function due to accumulation of mtDNA deletions but show no signs of premature aging. They represent a previously uncharacterized mouse model for an adult-onset progressive mitochondrial disease.
Materials and Methods
Generation of Transgenic Twinkle Mice. This work was approved by the National Public Health Institute animal care committee, and all experiments were done in accordance with good practice of handling laboratory animals.
We designed targeting constructs to carry the substitution of threonine for alanine at the amino acid 360 of mouse Twinkle protein (NP_722491) (A360T, TwinkleAT mice hereafter), or an in-frame duplication of amino acids 353–365 (dup353–365, Twinkledup). The production of wild-type Twinkle overexpressor mice (TwinkleWT+), serving as controls for possible quantitative effects of Twinkle expression, has been described in ref. 12. The mouse Twinkle cDNA and intron 4 was cloned into pHBApr-1-neo as described in ref. 12. The inclusion of the fourth intron allowed the expression of full-length Twinkle as well as its alternative splice variant Twinky. The p.A360T amino acid change corresponding to human p.A359T missense mutation was introduced with site-directed mutagenesis. To produce the dup353–365 mutation, a silent point mutation 1077C→G was introduced to form the restriction site Pfl23II, which was used to insert the 39-bp duplicated sequence with oligonucleotides. The primer sequences used are available upon request. The final DNA constructs were linearized with BglII and NdeI and used in pronuclear microinjection of FVB/N mouse fertilized oocytes. Genotyping of the mice and the transgene expression level determination were done as described in ref. 12.
Morphologic Analysis. Tissue samples were frozen in isopentane/liquid nitrogen. Cryostat sections from various tissue samples were stained for simultaneous cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activity. For plastic embedding, muscle samples were fixed in 2.5% glutaraldehyde for 1.5 h, treated with 1% osmium tetroxide, dehydrated in ethanol, and embedded in epoxy resin. Semithin (1 μm) sections were stained with methyl blue (0.5% wt/vol) and boric acid (1% wt/vol). For transmission electron microscopy (JEOL 1200EX electron microscope), ultrathin (60–90 nm) sections were cut on grids and stained with uranyl acetate and lead citrate.
Southern Blot. Mouse mtDNA quantitation was done as described in ref. 12.
Long PCR. Mouse mtDNA was amplified from 50 ng of total DNA with the primers 1953–1924 and 2473–2505 of mouse mtDNA (Expand Long Template PCR System, Roche) by using buffer 2 and PCR conditions: 92°C for 10 s and 68°C for 12 min, 30 cycles. PCR products were cloned either directly or after gel-purification (Gel Extraction Kit, Qiagen) to TOPO-TA vector (Invitrogen) and sequenced with vector primers and oligos corresponding to np 15 815-15 834, 315-334, and 16 024-16 005 of mouse mtDNA.
Laser-Capture Microdissection. Frozen sections (12 μm) were COX/SDH activity stained, dehydrated, and air dried. Individual skeletal muscle fibers were microdissected by using AutoPix Automated laser-capture microscope (Arcturus). Laser spots of 10 μm in diameter, a pulse power of 80mW, and a pulse width of 1500 μs were used. The fiber section was captured on CapSure cap (Arcturus), and the portion of the transfer film containing the fiber was cut out under microscope. The samples were digested in 20 μl of 10 mM Tris·HCl, pH 8.1/0.4 mg/ml proteinase K at 65°C for 3 h. The deletion molecules were amplified with oligos 2510–2529 and 15678–15659 of mouse mtDNA by using Dynazyme polymerase (Finnzymes) (95°C for 60 s, 56°C for 60 s, and 72°C for 60 s, 35 cycles).
Analysis of mtDNA Point Mutations. Total DNA was extracted from the quadriceps femoris muscle of 18-month-old mice. Somatic mtDNA mutation loads of two Twinkledup, one TwinkleWT+, and two control mice were determined by PCR, cloning, and sequencing as in ref. 19 with primers specific for cytochrome b gene (np 14 073-14 906) and the noncoding control region (15 357-15 138) of mouse mtDNA. More than 31,000 nucleotides of each region were analyzed from all samples.
Respiratory Chain Enzyme Activity Measurements. Mitochondria were isolated from quadriceps femoris, tibialis anterior, biceps femoris, and triceps surae muscles from >12-month-old Twinkledup mice (n = 4) and age-matched controls (n = 5). The respiratory chain enzyme activities of complexes I and III (NADH:cytochrome c reductase), complexes II and III (succinate:cytochrome c reductase), complex II (succinate dehydrogenase), complex IV (cytochrome c oxidase), and citrate synthase were determined as described in ref. 20. The oxygen consumption of mitochondria was determined with pyruvate plus malate, succinate plus rotenone, ascorbate plus TMPD, palmitoyl CoA, and α-ketoglutarate as substrates.
Functional Testing. Voluntary cage-wheel running. Hamster-sized metal cage wheels (diameter 14.5 cm) with digital magnetic counters (BC 800 Sigma sport) were placed into 47 × 26 × 14.5-cm cages to measure the voluntary daily running distance, daily running time, maximum running speed, and average running speed of the mice for 5 weeks (21). Three Twinkledup mice, 8 TwinkleAT mice, and 10 littermates, all >12 months of age, were tested.
Grip strength. Maximal grip strength was measured with a grip strength analyzer (TSE Systems) of each mouse in the beginning of the study, and after the 5-week exercise program, recording the best of three trials as maximum strength.
Rotarod. The Rotarod test (Ugo Basile, Biological Research Apparatus) was performed with a starting speed of 6 rpm. If the mouse stayed on the bar for 10 s, the rotation accelerated. The time of falling and the best of three consecutive attempts was recorded. Eight Twinkledup mice, 15 TwinkleAT mice, and 19 littermates were tested for Rotarod performance and grip strength.
Statistical analysis. Unpaired t test was used in statistical analysis, and the data are presented as mean ± standard deviation.
Results
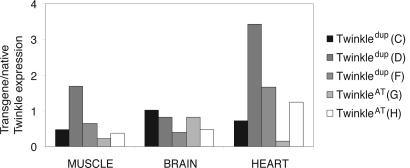
Generation of the Twinkle Transgenic Mice. We generated nine transgenic founder mice expressing mutant mouse Twinkle cDNA with either p.A360T change or an in-frame duplication of amino acids 353–365 under the ubiquitous β-actin promoter. The founders were used to produce transgenic mouse lines: two TwinkleWT+ (A and B), four Twinkledup (C, D, E, and F) and three TwinkleAT (G, H, and I) F1 lines. The distribution of genotypes in the litters followed Mendelian ratios. The F1 mice of lines A, C, and G were bred to produce the F2 mice homozygous for the transgene. The F1 lines had one to 12 transgene copies as determined by semiquantitative Southern blot (data not shown). The transgene expression levels in the muscle were low in the lines Twinkledup (E) and TwinkleAT (I), and they were excluded from further analysis. Other mutant lines displayed transgene expression levels from 0.2-fold to 3.4-fold compared to the endogenous Twinkle (Fig. 1), demonstrating that the insertion site influenced the β-actin promoter and the transgene expression. In general, the ratio of the transgene versus native allele expression in the mutant lines was close to the situation in patients with dominant disease, 1:1. The transgene-derived expression could not be confirmed at the protein level due to the lack of a specific antibody.
Fig. 1.
Transgene expression levels in the muscle, heart, and brain of the three transgenic Twinkledup (C, D, and F) and two TwinkleAT (G, H) mouse lines. The transgene expression levels were correlated with the expression of the native Twinkle gene.
Twinkle Transgenic Mice Show Characteristic Histological Features of Late-Onset Mitochondrial Myopathy. The overexpression of mutant Twinkle did not affect the mouse development or lifespan. The body weight of transgenic mice did not significantly differ from control littermates at any age.
Muscle Phenotype. No signs of mitochondrial dysfunction were noted during the first year of life. The 12-month-old Twinkledup mice showed COX negative (COX–, <1% of all fibers) muscle fibers in the histochemical analysis. The early COX– fibers did not show increased SDH activity (SDH+), indicating respiratory chain dysfunction without mitochondrial proliferation (Fig. 2b). Eighteen-month-old Twinkledup mice exhibited both COX– and COX–SDH+ fibers, suggesting that COX-deficiency preceded mitochondrial proliferation (Fig. 2d). The percentage of COX– fibers increased gradually with age: average percentage at the age of 18–24 months was 11.8 ± 4.4%, whereas it was 0.03 ± 0.01% in the age-matched controls (mean ± SD, P = 0.0008). All three F1 Twinkledup mouse lines (C, D, and F) presented similar levels of COX– fibers. TwinkleAT mice presented a mild muscle phenotype because only a few COX– fibers were found. As TwinkleWT+ mice or the nontransgenic littermates had virtually no COX– fibers even at the age of 22 months (Fig. 2 a and c), COX– fibers were not a result of the normal aging process or overexpression of the mitochondrial protein per se, but a consequence of the transgene.
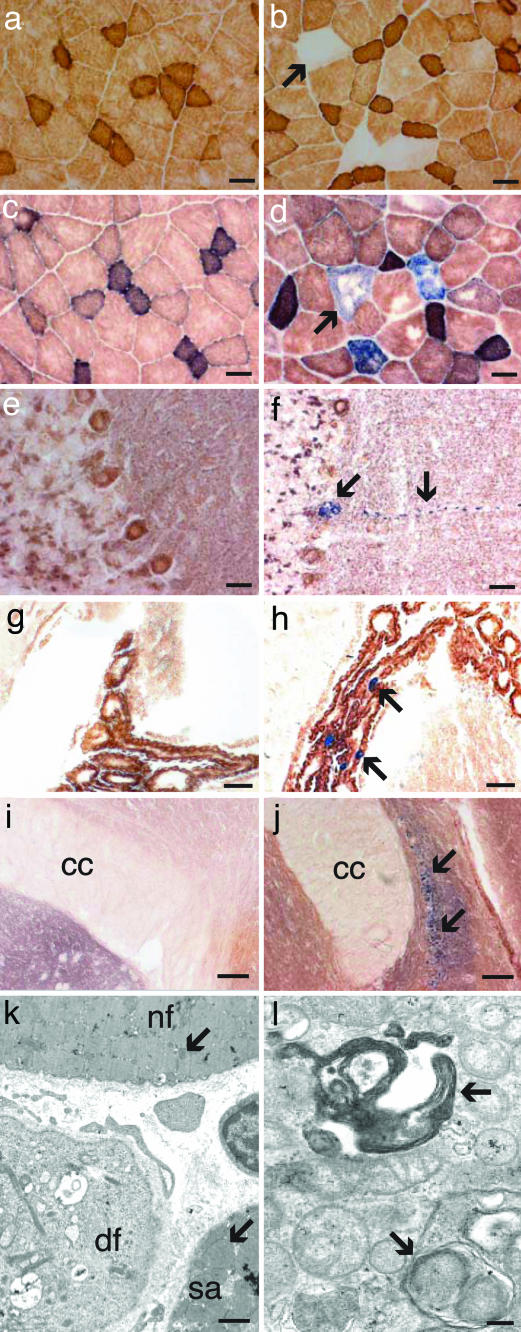
Fig. 2.
Cytochrome c oxidase activity of muscle and brain and the skeletal muscle ultrastructure. Double staining for COX/SDH activities of the quadriceps femoris of 12-month-old control (a) and Twinkledup mouse (b), the quadriceps femoris of 18-month-old control (c) and Twinkledup (d) mouse, cerebellum of 18-month-old control (e) and Twinkledup (f) mouse, choroid plexus of 18-month-old control (g) and Twinkledup (h) mouse, and indusium griseum of 18-month-old control (i) and Twinkledup (j) mouse (cc, corpus callosum). The arrows point to the affected cells of the transgenic mice. (k and l) Electron micrographs of ultrathin section of the quadriceps femoris of an 18-month-old Twinkledup mouse. (k) Normal-sized mitochondrion is pointed out by the arrow in the normal fiber (nf) and an enlarged mitochondrion in the fiber with subsarcolemmal accumulation of mitochondria (sa). In the diseased fiber (df), muscle fibrils have been replaced by abnormal large mitochondria with inclusions, peripheral cristae, and vacuoles. (l) The autophagosomes with mitochondria found from the diseased fibers are shown by arrows. (Scale bars: 25 μm, a–h; 100 μm, i and j; 1 μm, k; 200 nm, l.)
In electron microscopy of the Twinkledup mouse muscle, several fibers had increased the number and size of mitochondria (Fig. 2k). In mildly affected fibers, the mitochondria were enlarged, and their number had increased in the subsarcolemmal region, but they still had recognizable cristae structures. In the severely affected fibers, however, the myofibrillar structure had been replaced by large mitochondria with concentric cristae, resembling onion-like structures, and electron-dense inclusions. These changes were identical to those in the muscle of PEO patients with Twinkle defects, with the exception that typical paracrystalline inclusions were not identified. Mitochondria were seen frequently within autophagosomes, indicating mitochondrial degradation (Fig. 2l).
The heart muscle sections of aging mutant and control mice had roughly equal levels of COX–SDH+ fibers, similar to what has previously been reported for aging mice, indicating that our transgene did not substantially affect the heart muscle.
Biochemical analysis of the respiratory chain enzymes of a 20-month-old Twinkledup F2 mouse muscle, with the highest percentage of COX– fibers, showed moderately lowered COX activity and COX-dependent oxygen consumption with ascorbate as the substrate (50% and 64% of controls' mean, respectively, correlated against the nuclear-encoded complex II) (see supporting information, which is published on the PNAS web site). In many of the 20-month-old Twinkledup F1 mice, however, no significant reduction in the activities of the respiratory chain enzymes or oxygen consumption was detected, suggesting that the COX deficiency of individual muscle fibers was local and did not impair the total respiration capacity of the muscle.
Deficiency of COX in Distinct Neuronal Populations. Frozen brain sections of 18-month-old Twinkledup mice showed that ≈1% of Purkinje cells in the cerebellum were COX–SDH+ in both C and F lines, with COX deficiency and mitochondrial proliferation in the neuronal soma and dendrite (Fig. 2f). Other cerebellar cells had normal COX activity. In the cerebrum, large pyramidal neurons in the hippocampal CA2 region were mostly COX–SDH+, as well as neurons of indusium griseum, the region from which hippocampal cells developmentally derive (Fig. 2j). A few COX–SDH+ neurons were also identified from the olfactory bulbs, substantia nigra, and the hypothalamus (data not shown). Many of the metabolically active and mitochondria-rich epithelial secretory cells of the choroid plexus were also COX–SDH+ (Fig. 2h). No COX–SDH+ neurons or other brain cells were found from age-matched control littermates or TwinkleWT+ mice (Fig. 2 e, g, and i). The COX– regions could reflect either β-actin expression pattern or sensitivity of the affected cells to mitochondrial dysfunction. The regions with the highest endogeneous β-actin expression in mouse brain are the olfactory bulbs, motor cortex, and choroid plexus (22), of which the first and third showed evident COX– deficiency in our mice, whereas no COX– neurons were identified in the pyramidal neurons of the motor cortex.
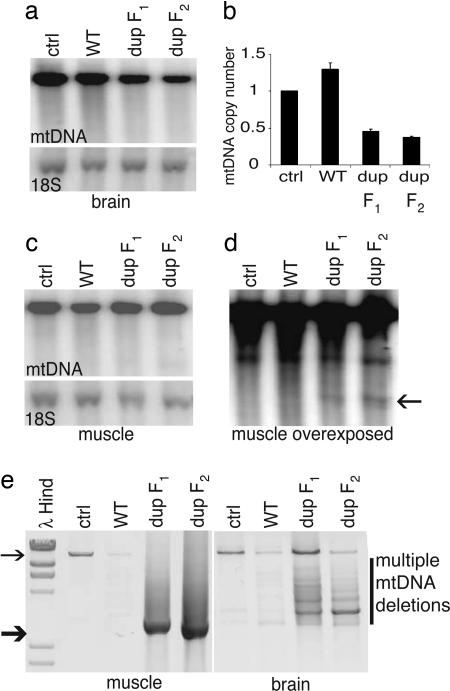
Mutant Mice Accumulate Mitochondrial DNA Deletions. We searched for multiple mtDNA deletions in our Twinkledup and control mice by long PCR and by Southern hybridization analysis, because these types of mutations accumulate in Twinkle-PEO patients. Amplification of brain mtDNA of Twinkledup mice resulted in multiple products, corresponding to full-size mtDNA and molecules with deletions of multiple sizes (Fig. 3e). Only the 16-kb product was amplifiable from the control brain. The similar deletion pattern of the brain mtDNA in different mice indicated deletion hotspots. However, from the muscle, only a single product of ≈3 kb was readily amplified, whereas the 16-kb product from the wild-type mtDNA amplified from all of the control samples (Fig. 3e). DNA sequence analysis revealed that this 3-kb product represented multiple different deletion molecules, which all had their 5′ breakpoints between 2707–3020 and 3′ breakpoints between 15376–15441 (supporting information). Only 12S rRNA, 16S rRNA, and the noncoding region of mtDNA remained in these molecules (Fig. 4a). The deletions could not be seen on Southern blots (Fig. 3 a and c), except for the 3-kb mutant population in the muscle, seen in the samples of Twinkledup F1 and F2 mice upon overexposure of autoradiography, estimated to constitute ≈5% of total mtDNA in the muscle (Fig. 3d). We used laser-capture microdissection to obtain DNA from single muscle fibers and were able to correlate the deletion molecules with some COX– fibers and not any of the COX+ fibers by single-fiber PCR (data not shown). The heart DNA of aging mice, mutated or control, contained multiple mtDNA deletions both by long PCR and overexposed Southern blot, but they were not in excess in the mutant mouse heart (data not shown). Quantification of mtDNA copy number against the nuclear 18S rRNA gene in Twinkledup mouse tissues revealed a clear mtDNA depletion in the brain (46% of control value in dup F1 mice and 37% in dup F2 mice, P < 0.0001 for both; Fig. 3b). In the muscle and the heart of the Twinkledup mice, mtDNA levels were similar to those of the nontransgenic littermates.
Fig. 3.
Mitochondrial DNA analysis and copy number determination. (a and c) Southern blots of muscle and brain DNA of 22-month-old control, TwinkleWT+ mice, and Twinkledup F1 mouse and an 18-month-old Twinkledup F2 mice probed with mtDNA, and nuclear 18S rRNA gene. (b) Quantitation of brain mtDNA copy number of the mice against the nuclear 18S rRNA gene. (d) Overexposed Southern blot of muscle DNA shown in c. The 3-kb band found from Twinkledup mice is pointed by the arrow. (e) Long PCR of muscle and brain DNA of the same mice. The full-length mtDNA is indicated by the thin arrow and the most prevalent deletion molecule size of muscle mtDNA (≈3 kb) by the thick arrow.
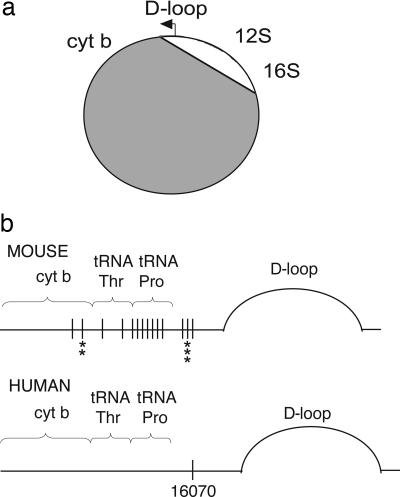
Fig. 4.
mtDNA deletions of the muscle and brain of the Twinkledup mouse, determined by long PCR, cloning, and DNA sequencing. (a) Schematic diagram of mouse mtDNA showing the deleted segment in gray (see supporting information for details). Only the D-loop, 12S rRNA, and 16S rRNA were commonly spared. (b) Schematic presentation of the mouse and human mtDNA region harboring the 5′ deletion breakpoints. Black vertical lines show the 5′ deletion breakpoints identified in this study, and the asterisks below indicate how often the same 5′ breakpoint was found from dissimilar deletion molecules. The common 5′deletion breakpoint of human multiple mtDNA deletions (16070) is indicated.
Our 18-month-old Twinkledup mice did not have an increased somatic mtDNA point mutation load in their muscle samples: Only 0.45 mutations per 10 kb in the cytochrome b region and 0.25 mutations in the control region were present, when >33 kb of cloned mtDNA sequence was analyzed per mouse.
The Physical Performance of the Mice Is Not Compromised. Because of the marked histological changes in the muscle, we studied the muscle strength and performance of our mice. The rotating bar, Rotarod, performance reflects the ability to sustain complex coordinated movements; the test of Twinkledup and TwinkleAT mice showed no statistical significant difference compared to the littermates (supporting information). Likewise, the maximal grip strength was not affected by the muscle changes and was similar to the control littermates. The distance run on voluntary running-wheel exercise has been shown to reflect the endurance capacity. We found no signs of significant muscle weakness or impairment of exercise capacity in Twinkledup or TwinkleAT mice, but the daily running duration and distance of the transgenic mice was higher compared to controls (P = 0.05). The distance per day for each week (mean ± SD) was 1.3 ± 0.5 km in controls, 2.6 ± 0.6 km in mice harboring the TwinkleAT mutation, and 3.7 ± 0.7 km in the Twinkledup mice (supporting information).
Discussion
We report here a mouse model for an adult-onset mitochondrial disease. Our transgenic mice, expressing the mitochondrial Twinkle helicase with dominant PEO mutations, replicated faithfully the key histological and molecular findings of the human PEO disease. This model developed a morphologically clear mitochondrial myopathy but affected the well being of the mouse only mildly, thus offering great promise for evaluation of therapeutic strategies.
The Twinkle mutations expressed in the mice corresponded to two PEO patient mutations, dup352–364 and A359T. The inframe duplication is the structurally most severe mutation so far described in Twinkle. The A359T mutation was chosen, because in bacteriophage T7, the analogous mutation A257T was a gain-of-function mutation, resulting in enhanced dTTPase activity of the helicase (23). The patients with dup352–364 presented with autosomal dominant PEO, generalized muscle weakness, avoidant personality traits, and major depression with the age of onset between 20 to 51 years (8, 9). The missense mutation A359T in human patients caused late-onset PEO at ≈50 years with moderate myopathy and no CNS involvement (M. Zeviani, personal communication). Our Twinkledup mice developed the first histological signs of mitochondrial myopathy at the age of 12 months, mimicking the relative age of onset of human disease when related to the expected lifespan. The myopathy of the mice progressed, as judged by the growing severity of the morphological changes. The amount of COX– muscle fibers in PEO patients with dup352–364 was 2–11% (9) and in Twinkledup mice was ≈12%. The three independent Twinkledup transgenic lines developed the same muscle phenotype showing that the muscle changes were caused by the mutant Twinkle and not by the transgene insertion site. The TwinkleAT mice with the homologous mutation presented a slow progression of COX deficiency compared to the Twinkledup mice. The overall mouse muscle phenotype closely mimics the findings in the PEO patients.
A hallmark of the PEO disease is the accumulation of multiple large mtDNA deletions in the patients' postmitotic tissues, the mutant mtDNA reaching up to 60% of total mtDNA in some brain regions and 40% in the skeletal muscle (1, 24). We observed accumulation of similar deletions in the corresponding mouse tissues. The number of deletion molecules increased with age, along with the progressive respiratory chain dysfunction, judged by the increase of the COX-deficient fibers. However, the amount of deleted mtDNA in the mice was quite low, resembling the findings in juvenile PEO patients with dup352–364 mutation (9) and in patients with POLG-related ataxias (4). The low mutant mtDNA amount may reflect the short lifespan of mice: in humans mtDNA deletions became visible in Southern blot at only ≈20 years of age, whereas our mice were maximally ≈2 years when killed. Interestingly, the low number of mutant mtDNA did not reflect the quantity of COX-deficient muscle fibers, because the number of abnormal fibers was similar in PEO patients and our mice. Our mice had the somatic point mutation load and total mtDNA copy number similar to controls in their muscle and, therefore, these factors did not exacerbate COX deficiency. The COX deficiency seemed to clearly precede the mitochondrial accumulation, because 1-year-old mice had some completely COX– fibers, but SDH+ fibers were first observed in 18-month-old mice.
The muscle atrophy and mitochondrial dysfunction caused surprisingly little functional consequences in our mice. Biochemical analysis showed mildly decreased or low normal activities of the respiratory chain enzyme complexes I, III, and IV. These complexes contain mtDNA-encoded subunits, and the finding strongly supports the causal relationship of mtDNA defects and respiratory chain deficiency. Similar activities are typical for PEO patients with dup352–364 (9), reflecting the low proportion of affected fibers in the muscle homogenate. The physical performance, exercise capacity and muscle strength, of our mice was good. Actually, the mutant mice seemed to exercise voluntarily slightly more than their wild-type littermates. It is tempting to speculate that the increased urge to run would be a consequence of the transgene: a result of direct brain involvement or spontaneous behavior to compensate for the muscle atrophy. However, these conclusions require further studies with a large number of mice with different genetic background. We show, however, that mouse muscle tolerates a considerable number of dysfunctional fibers until its function becomes compromised. This phenotype agrees well with the findings in Twinkledup patients, who often have considerable subjective exercise intolerance but mild objective muscle weakness (25).
To examine the deletion mechanisms, we characterized in detail the mtDNA deletion breakpoints in the brain and muscle of our mice. The Twinkledup mouse muscle harbored mutant mtDNA, the majority of which carried a ≈13-kb nonclonal mtDNA deletion. The products all consisted only of rRNA genes and the D-loop region but had individual 5′ and 3′ breakpoint sequences at close-by sites. These breakpoints either contained no or short direct repeats of 4–5 bp, consistent with class II deletions. These 3-kb “minimal mtDNA molecules” closely resembled the 3.75-kb “common sublimons” previously found in the control human heart (26). The brain mtDNA also resulted in a range of amplification products from full-length to ≈3 kb in size. This deletion pattern closely resembled the findings in human patients (24). Upon breakpoint analysis, we identified similar deletion hotspots in the brain as in the muscle, suggesting that these sites form a major deletion hotspot in Twinkle mice. Most importantly, the 3′ deletion breakpoints, bp 15180–15441 of mouse mtDNA, overlapped with the human base pair 16070, a previously described breakpoint hotspot of PEO-Twinkle patients (27) and the common sublimon (26). Furthermore, a recent study targeted PstI restriction enzyme to the mouse skeletal muscle mitochondria, resulting in continuous generation of double-strand breaks. These mice had 3′ PstI-independent deletion breakpoints at bp 15368–15439 (28), overlapping with our breakpoints. The hotspot region in human and mouse mtDNA is located between tRNAPro and the D-loop and contains the most highly divergent primary sequence of mtDNA (29). It seems evident that the general mechanism causing multiple deletions with breakpoints at the specific site is not due to the primary sequence itself but its secondary structure or functional location. The PstI mice strongly suggested that double-strand breaks mediate the formation of class II mtDNA deletions in mammals (28). The intriguing hypothesis arising from the present and previous data are that the primary consequence of Twinkledup defect was generation of double-strand breaks, as we have suggested based on deletion breakpoint analysis of PEO-Twinkle patients (27). Furthermore, we suggest a specific conserved role for the hotspot region in double-strand break repair or as a recombination site for free 5′ ends of the double-strand breaks.
POLG and Twinkle defects have recently been found to underlie a wide phenotype range expanding to neurodegenerative diseases (4, 5). Therefore, we carefully looked for CNS involvement in two different Twinkledup mouse lines. We identified populations of large neurons that were COX–SDH+ in both lines: cerebellar Purkinje cells, hippocampal CA2 pyramidal neurons, and those of indusium griseum. The COX deficiency in these neurons may result from mtDNA depletion or accumulation of deleted mtDNA in selected cells. COX-deficient Purkinje cells have not been previously reported in mice or in humans. Loss of Purkinje cells was noted in an autopsy of PEO patient with dup352–364 mutation and in mtDNA maintenance defects, such as mitochondrial spinocerebellar ataxia and Alpers syndrome (4, 6, 24). Notably, the COX–SDH+ hippocampal neurons in our mice resembled closely those reported in aged humans and in Alzheimer patients (30, 31). In our mice, the neuronal COX deficiency did not directly follow the expression pattern of mouse β-actin, and the deficiency may manifest in those neuronal populations that are most susceptible to defects in mtDNA maintenance and mitochondrial dysfunction. Thus, our mouse model provides a promising tool to understand neuronal cell type-specific mtDNA maintenance.
MtDNA mutations have recently been linked to premature aging because the mice with proofreading-deficient POLG were found to have large numbers of somatic mtDNA point mutations already at the age of two months and to present phenotypes associated with aging in mammals (7). The “mutator mice” were reported to also have deleted, linear mtDNA, the amount of which did not increase over time. Twinkle is the partner of POLG in replication, and it was of interest to notice that our deletor mice did not age prematurely, although they accumulate mtDNA deletions, leading to progressive respiratory chain deficiency. Our data suggest that premature aging might require extensive mtDNA mutagenesis, or alternatively, the phenotype of the mutator mice is due to a specific function of POLG protein not associated with mtDNA mutagenesis.
The Twinkledup mice are mtDNA disease models that replicate the manifestations of a specific patient mutation. Previous mouse models for mitochondrial respiratory dysfunction include gene knockouts of essential proteins for mtDNA maintenance: inactivation of adenine-nucleotide translocator-1 resulted in myopathy and cardiac hypertrophy at 4–6 months of age (15). Tissue-specific knockouts of Tfam elucidated the consequences of the loss of mitochondrial transcripts and severe respiratory chain deficiency in specific cell types (18). The ΔmtDNA mice had high levels of a single mtDNA deletion in most tissues already at birth (17), resulting in respiratory chain deficiency in heart, skeletal muscle, and kidney and making it a good model for an early onset multisystem mitochondrial disease. The ΔmtDNA mice died at 6 months of age due to renal failure. Mice expressing PstI in mitochondria developed mitochondrial myopathy at 7 months of age but could not reproduce (28). In contrast to all previous models, the Twinkledup mice specifically model a late-onset progressive mitochondrial disease without compromising lifespan or reproduction.
Therapy trials for mitochondrial disorders are exceptionally challenging, because large homogeneous patient materials allowing double-blind test setups are not available even in large centers. Our transgenic Twinkledup mouse model enables studies of long- and short-term effects of exercise or different therapeutic ideas in a double-blinded setting, with genetically homogeneous and large study material. It enables follow-up of presymptomatic disease progression and intervention, as well as tests directed to reveal the pathogenesis of a late-onset disease. These deletor mice are an ideal tool to study mtDNA disease.
Supplementary Material
Acknowledgments
We thank the staff from the animal units of National Public Health Institute and Haartman Institute for skillful assistance. This work was supported by Helsinki Biomedical Graduate School (H.T.); Academy of Finland Centre of Excellence program (A.S. and J.N.S.); Sigrid Juselius Foundation and Helsinki University research funds (to A.S.); and the Medical Research Fund of Tampere University Hospital and the European Community's Sixth Framework Program for Research Priority 1 “Life Sciences, Genomics, and Biotechnology for Health” Contract LSHM-CT-2004-503116 (to J.N.S.).
Author contributions: H.T., K.P.M., A.J., J.N.S., and A.S. designed research; H.T., K.P.M., S.W., I.L., E.Y., and A.P. performed research; H.T., K.P.M., S.W., I.L., J.N.S., A.P., and A.S. analyzed data; and H.T., A.J., J.N.S., and A.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: COX, cytochrome c oxidase; PEO, progressive external ophthalmoplegia; POLG, polymerase γ; SDH, succinate dehydrogenase.
References
- 1.Zeviani, M., Servidei, S., Gellera, C., Bertini, E., DiMauro, S. & DiDonato, S. (1989) Nature 339, 309–311. [DOI] [PubMed] [Google Scholar]
- 2.Moraes, C. T., Shanske, S., Tritschler, H. J., Aprille, J. R., Andreetta, F., Bonilla, E., Schon, E. A. & DiMauro, S. (1991) Am. J. Hum. Genet. 48, 492–501. [PMC free article] [PubMed] [Google Scholar]
- 3.Van Goethem, G., Dermaut, B., Lofgren, A., Martin, J. J. & Van Broeckhoven, C. (2001) Nat. Genet. 28, 211–212. [DOI] [PubMed] [Google Scholar]
- 4.Van Goethem, G., Luoma, P., Rantamaki, M., Al Memar, A., Kaakkola, S., Hackman, P., Krahe, R., Lofgren, A., Martin, J. J., De Jonghe, P., et al. (2004) Neurology 63, 1251–1257. [DOI] [PubMed] [Google Scholar]
- 5.Luoma, P., Melberg, A., Rinne, J. O., Kaukonen, J. A., Nupponen, N. N., Chalmers, R. M., Oldfors, A., Rautakorpi, I., Peltonen, L., Majamaa, K. et al. (2004) Lancet 364, 875–882. [DOI] [PubMed] [Google Scholar]
- 6.Naviaux, R. K. & Nguyen, K. V. (2004) Ann. Neurol. 55, 706–712. [DOI] [PubMed] [Google Scholar]
- 7.Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A. T., Bruder, C. E., Bohlooly, Y. M., Gidlof, S., Oldfors, A., Wibom, R. et al. (2004) Nature 429, 417–423. [DOI] [PubMed] [Google Scholar]
- 8.Spelbrink, J. N., Li, F. Y., Tiranti, V., Nikali, K., Yuan, Q. P., Tariq, M., Wanrooij, S., Garrido, N., Comi, G., Morandi, L. et al. (2001) Nat. Genet. 28, 223–231. [DOI] [PubMed] [Google Scholar]
- 9.Suomalainen, A., Majander, A., Wallin, M., Setala, K., Kontula, K., Leinonen, H., Salmi, T., Paetau, A., Haltia, M., Valanne, L. et al. (1997) Neurology 48, 1244–1253. [DOI] [PubMed] [Google Scholar]
- 10.Lewis, S., Hutchison, W., Thyagarajan, D. & Dahl, H. H. (2002) J. Neurol. Sci. 201, 39–44. [DOI] [PubMed] [Google Scholar]
- 11.Korhonen, J. A., Pham, X. H., Pellegrini, M. & Falkenberg, M. (2004) EMBO J. 23, 2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyynismaa, H., Sembongi, H., Bokori-Brown, M., Granycome, C., Ashley, N., Poulton, J., Jalanko, A., Spelbrink, J. N., Holt, I. J. & Suomalainen, A. (2004) Hum. Mol. Genet. 13, 3219–3227. [DOI] [PubMed] [Google Scholar]
- 13.Sawaya, M. R., Guo, S., Tabor, S., Richardson, C. C. & Ellenberger, T. (1999) Cell 99, 167–177. [DOI] [PubMed] [Google Scholar]
- 14.Singleton, M. R., Sawaya, M. R., Ellenberger, T. & Wigley, D. B. (2000) Cell 101, 589–600. [DOI] [PubMed] [Google Scholar]
- 15.Graham, B. H., Waymire, K. G., Cottrell, B., Trounce, I. A., MacGregor, G. R. & Wallace, D. C. (1997) Nat. Genet. 16, 226–234. [DOI] [PubMed] [Google Scholar]
- 16.Huo, L. & Scarpulla, R. C. (2001) Mol. Cell. Biol. 21, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue, K., Nakada, K., Ogura, A., Isobe, K., Goto, Y., Nonaka, I. & Hayashi, J. I. (2000) Nat. Genet. 26, 176–181. [DOI] [PubMed] [Google Scholar]
- 18.Larsson, N. G. & Rustin, P. (2001) Trends Mol. Med. 7, 578–581. [DOI] [PubMed] [Google Scholar]
- 19.Spelbrink, J. N., Toivonen, J. M., Hakkaart, G. A., Kurkela, J. M., Cooper, H. M., Lehtinen, S. K., Lecrenier, N., Back, J. W., Speijer, D., Foury, F. et al. (2000) J. Biol. Chem. 275, 24818–24828. [DOI] [PubMed] [Google Scholar]
- 20.Majander, A., Rapola, J., Sariola, H., Suomalainen, A., Pohjavuori, M. & Pihko, H. (1995) J. Neurol. Sci. 134, 95–102. [DOI] [PubMed] [Google Scholar]
- 21.Allen, D. L., Harrison, B. C., Maass, A., Bell, M. L., Byrnes, W. C. & Leinwand, L. A. (2001) J. Appl. Physiol. 90, 1900–1908. [DOI] [PubMed] [Google Scholar]
- 22.Zapala, M. A., Hovatta, I., Ellison, J. A., Wodicka, L., Del Rio, J. A., Tennant, R., Tynan, W., Broide, R. S., Helton, R., Stoveken, B. S., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 10357–10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washington, M. T., Rosenberg, A. H., Griffin, K., Studier, F. W. & Patel, S. S. (1996) J. Biol. Chem. 271, 26825–26834. [DOI] [PubMed] [Google Scholar]
- 24.Suomalainen, A., Majander, A., Haltia, M., Somer, H., Lonnqvist, J., Savontaus, M. L. & Peltonen, L. (1992) J. Clin. Invest. 90, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lofberg, M., Lindholm, H., Naveri, H., Majander, A., Suomalainen, A., Paetau, A., Sovijarvi, A., Harkonen, M. & Somer, H. (2001) Neuromuscul. Disord. 11, 370–375. [DOI] [PubMed] [Google Scholar]
- 26.Kajander, O. A., Rovio, A. T., Majamaa, K., Poulton, J., Spelbrink, J. N., Holt, I. J., Karhunen, P. J. & Jacobs, H. T. (2000) Hum. Mol. Genet. 9, 2821–2835. [DOI] [PubMed] [Google Scholar]
- 27.Wanrooij, S., Luoma, P., van Goethem, G., van Broeckhoven, C., Suomalainen, A. & Spelbrink, J. N. (2004) Nucleic Acids Res. 32, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava, S. & Moraes, C. T. (2005) Hum. Mol. Genet. 14, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walberg, M. W. & Clayton, D. A. (1981) Nucleic Acids Res. 9, 5411–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cottrell, D. A., Blakely, E. L., Johnson, M. A., Ince, P. G., Borthwick, G. M. & Turnbull, D. M. (2001) Neurobiol. Aging 22, 265–272. [DOI] [PubMed] [Google Scholar]
- 31.Cottrell, D. A., Blakely, E. L., Johnson, M. A., Ince, P. G. & Turnbull, D. M. (2001) Neurology 57, 260–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.