Abstract
Although pneumonic plague is the deadliest manifestation of disease caused by the bacterium Yersinia pestis, there is surprisingly little information on the cellular and molecular mechanisms responsible for Y. pestis-triggered pathology in the lung. Therefore, to understand the progression of this unique disease, we characterized an intranasal mouse model of primary pneumonic plague. Mice succumbed to a purulent multifocal severe exudative bronchopneumonia that closely resembles the disease observed in humans. Analyses revealed a strikingly biphasic syndrome, in which the infection begins with an antiinflammatory state in the first 24-36 h that rapidly progresses to a highly proinflammatory state by 48 h and death by 3 days. To assess the adaptation of Y. pestis to a mammalian environment, we used DNA microarray technology to analyze the transcriptional responses of the bacteria during interaction with the mouse lung. Included among the genes up-regulated in vivo are those comprising the yop-ysc type III secretion system and genes contained within the chromosomal pigmentation locus, validating the use of this technology to identify loci essential to the virulence of Y. pestis.
Keywords: inflammation, microarray, Yersinia pestis
Pneumonic plague is the deadliest manifestation of disease caused by the bacterium Yersinia pestis. Although rare compared with the bubonic form of plague, which is acquired by skin penetration, primary pneumonic plague is highly contagious and almost always fatal. The current worldwide incidence of plague is low by historical standards, but the possible combination of widespread aerosol dissemination and rapid disease progression are of particular concern for defense against bioterrorism (1).
In cases of primary pneumonic plague in humans, microscopic examination of lung tissue reveals multiple histological patterns, including acute pneumonia, intraalveolar hemorrhage and edema, and the presence of extracellular bacteria in the alveoli but not the interstitium (2). In addition, extensive neutrophilic infiltrate and fibrin deposition have been observed, and in some cases a complete loss of recognizable alveolar architecture results from the infection (3, 4). Studies of experimental primary pneumonic plague in monkeys, mice, and guinea pigs showed similar pathologic effects, including extensive intraalveolar edema, massive bacterial proliferation in the small airways, and numerous neutrophils in the alveoli (5-8).
How Y. pestis triggers pulmonary pathology is largely unexplored, as are the bacterial responses to this dramatically changing host environment. DNA microarray technology has been widely used to assess transcriptional changes in bacterial gene expression in vitro, but analyses of the bacterial transcriptome during host infection have been hampered by two significant problems (9). These include the excess amounts of copurified eukaryotic RNA that may produce nonspecific hybridization signals on the microarray, and the oftenlimiting amounts of bacterial RNA extracted from an animal, which would otherwise require large numbers of animals for analysis. Progress has been made by multiple groups to overcome these problems (10-12), although different pathogens, animal hosts, and routes of infection necessitate empirically derived adaptations for each study. Nonetheless, advances in microarray technology will allow for more complete analyses of animal models of infection.
Although recent studies have examined the end result of pneumonic plague infection (i.e., death) in animal models, little is known about the early events in this disease. We used Y. pestis strain CO92, isolated from a fatal case of human pneumonic plague (3), in combination with an intranasal inbred mouse model of infection to more fully characterize the interaction between the bacterium and the host during primary pneumonic plague. In addition, we present data on the transcriptional expression profile of Y. pestis in the lungs compared with bacteria grown in vitro by use of DNA microarray technology.
Materials and Methods
Bacterial Strains and Culture Conditions. The virulent wild-type Y. pestis strain CO92 and its plasmid-cured derivative CO92 pCD1- were obtained from the U.S. Army, Ft. Detrick, MD. Y. pestis strain KIM6(pCD1Ap)+ (13) was a kind gift from Robert Perry (University of Kentucky, Lexington). The presence or absence of pCD1, pMT1, pPCP1, and the pgm locus was confirmed by PCR for each strain. Y. pestis was routinely grown on brain-heart infusion (BHI) agar (Difco) at 26°C for 2-3 days. For liquid cultures, Y. pestis was grown in BHI broth at 26°C for 6-8 h in a roller drum before being dilutedtoanOD620 of 0.05-0.1 in 10 ml of BHI broth with 2.5 mM CaCl2 in a 125-ml Erlenmeyer flask. Unless otherwise indicated, bacteria were incubated at 37°C in a water bath shaker set at 250 rpm for 16-18 h.
Animals. Animal experiments were approved by the Washington University Animal Studies Committee, protocol no. 20020257. Pathogen-free 6- to 8-week-old female C57BL/6 mice were obtained from The Jackson Laboratory and were housed in highefficiency particulate air-filtered barrier units kept inside biological safety cabinets for the duration of the experiments. Mice were given food and water ad libitum and were kept at 25°C with alternating 12-h periods of light and dark. Bacteria were grown in BHI broth, washed in sterile PBS, and maintained at 37°C. Mice were lightly anesthetized and inoculated by the intranasal route with 20 μl of Y. pestis in PBS. Numbers of colony-forming units (cfu) inoculated were determined by plating serial dilutions onto BHI agar. Animals that were clearly moribund or on the verge of death were killed with an overdose of pentobarbital sodium (150 mg/kg).
LD50, Kinetics, and Survival Curves. Groups of four to five mice were infected intranasally with serial 10-fold dilutions of Y. pestis strain CO92, ranging from 10 to 1 × 105 cfu. Mice were monitored twice daily for 7 days, and any surviving mice were killed with pentobarbital sodium (150 mg/kg). For experiments examining the kinetics of infection, groups of five mice were infected intranasally with 1 × 104 cfu of Y. pestis strains CO92 or CO92 pCD1-. At various times postinfection, mice were killed, the lungs and spleens surgically removed and homogenized in 0.5 ml of sterile PBS, and serial dilutions were plated onto BHI agar. Results are reported as cfu per organ.
Histopathology. Groups of five mice were infected intranasally with 1 × 104 cfu of Y. pestis strain CO92. Uninfected mice and mice infected for 24, 48, or 72 h were killed with pentobarbital sodium (150 mg/kg) and their lungs inflated with 10% neutral buffered formalin via cannulation of the trachea. Lungs and spleens were removed and fixed in 10% formalin overnight before being embedded in paraffin. Five-micrometer sections of tissue were stained either with hematoxylin/eosin or by Steiner (silver) stain before being examined.
Cytokine Analysis. Levels of IL-10, IL-6, Il-12p70, TNF, IFN-γ, and monocyte chemoattractant protein (MCP-1) present in mouse lungs were measured with the mouse inflammation cytometric bead array kit (BD Biosciences, Franklin Lakes, NJ). Groups of five mice were infected intranasally with 1 × 104 cfu of Y. pestis strain CO92. Lungs from uninfected mice and mice infected for 24, 48, or 72 h were removed and homogenized in 0.5 ml of ice-cold PBS containing protease inhibitors (Roche Diagnostics). Homogenates were centrifuged at 16,000 × g for 2 min, and the supernatants were passed through a 0.22-μm filter. Filtrates were analyzed as specified by the manufacturer. Cytokine concentrations were determined by using the cytometric bead array analysis software, Ver. 1.1.
RNA Isolation, T7-Based RNA Amplification, and Microarray Analysis. A Y. pestis gene-specific microarray, consisting of 70-mer oligonucleotides representing 100% of the ORFs of strain CO92, was constructed (Supporting Text, which is published as supporting information on the PNAS web site). Two independent identical experiments were conducted as follows: 10 mice were infected with Y. pestis strain CO92 and after 48 h, mice were killed, lungs were lavaged with PBS, and the resulting material was pooled. RNA was extracted from lavage-derived bacteria and from a culture of CO92 grown for 12 h at 37°C in BHI broth with 2.5 mM CaCl2 (Supporting Text). To create technical replicates for each biological replicate, RNAs were divided into two samples. These samples were independently amplified, labeled, and hybridized to the arrays in such a way that each of the two lavage samples was paired with each of the two culture-derived samples. Thus four technical replicates were tested, and each was dye-swapped for a total of eight DNA microarrays that were used for each experiment (Supporting Text). Slides were scanned immediately after hybridization and analyzed with genespring (Agilent Technologies, Palo Alto, CA). Within each experiment genes that had signal-to-control or control-to-signal ratios >2.0 (P < 0.05, Bonferroni Step-Down Multiple Testing correction) in either the high- or low-photomultiplier scans were considered to be significantly regulated (Supporting Text). Only genes that were significantly regulated in all the technical replicates were included in the final list (Table 2, which is published as supporting information on the PNAS web site).
Microarray Validation. A randomly selected group of genes (n = 27) that were differentially regulated as determined by microarray hybridization were analyzed by quantitative RT-PCR (qRT-PCR) (Supporting Text). Array data were considered valid if the fold change of each gene tested by qRT-PCR was >2.0 and in the same direction as determined by microarray analysis.
Results
Survival of C57BL/6 Mice Infected Intranasally with Y. pestis. The progression of primary pneumonic plague in humans and other animals is rapid; symptoms develop 24-48 h after exposure, followed by death within 3-4 days (6, 14). To better understand the course of primary pneumonic plague in the mouse model, we inoculated C57BL/6 mice by the intranasal route with increasing doses of Y. pestis from 10 to 1 × 105 cfu and monitored the development of symptoms and survival of the animals. After 24 h, all mice looked healthy and were behaving normally, with no visible signs of symptoms. By 36 h, however, mice receiving the highest dose of Y. pestis appeared listless, hunched up and huddled together, breathing rapidly, and had little reaction to being handled. By 48 h, mice receiving 1 × 104 cfu exhibited similar symptoms, and by 60 h, mice began to die from the infection (Fig. 4, which is published as supporting information on the PNAS web site). By 3.5 days, 100% of the mice receiving 105 or 104 cfu of Y. pestis had died, whereas five of nine mice exposed to 1 × 103 cfu were dead. Mice continued to succumb up to 4.5 days after inoculation, after which all remaining mice survived. As expected, the percentage of surviving mice depended on the dose of the inoculation, and there was a direct correlation between the development of symptoms and the progression to death (no mice that exhibited symptoms survived). The LD50 of Y. pestis strain CO92 administered intranasally for two experiments was calculated as 2.8 × 102 and 2.4 × 102 cfu, respectively. Mice infected with another strain of Y. pestis, KIM6(pCD1Ap)+, showed similar results to CO92 in this model (data not shown). Thus, for all future mouse experiments, we chose the lowest dose tested that consistently gave a 100% mortality rate, 1 × 104 cfu.
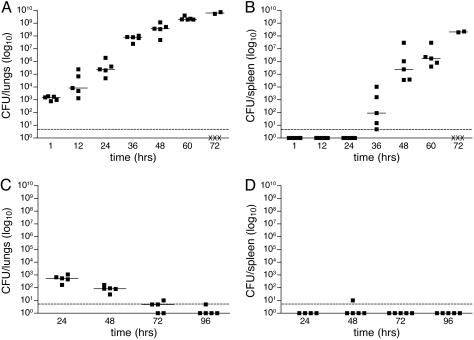
Colonization of Mouse Tissues After Intranasal Infection with Y. pestis. We assessed the kinetics of Y. pestis CO92 infection after intranasal inoculation by calculating the cfu present in the lungs and spleen at various times during infection. One hour postinoculation, ≈103 cfu, or 10% of the bacteria, were recovered from the lungs of the mice (Fig. 1A), which is similar to the percent present at early time points in other animal models (5, 15). Bacterial load proceeded to rise rapidly, reaching 1010 by day 3, with the greatest increase occurring between 24 and 36 h. This corresponds to the time in which Y. pestis appeared in the spleens (Fig. 1B), indicative of dissemination from the lungs into the peripheral tissues. Bacterial load in the spleens also continued to increase with time, approaching 109 cfu by 72 h. In contrast, an isogenic strain of CO92 lacking the pCD1 plasmid was unable to cause a lethal infection, was cleared from the lungs, and was not detected in the spleen (Fig. 1 C and D).
Fig. 1.
Kinetics of infection with Y. pestis strain CO92 or CO92 pCD1-. Bacteria (1 × 104) were introduced intranasally into C57BL/6 mice, and cfu per organ in the lungs (A and C) and spleen (B and D) were determined every 12 h for 3 days (CO92, A and B) or every 24 h for 4 days (CO92 pCD1-, C and D). The limit of detection is indicated by a dashed line. Each point indicates cfu recovered from a single animal. Symbols below the limit of detection represent mice that survived but did not have detectable numbers of bacteria; “X” indicates mice that succumbed to the infection. A solid line indicates the median of cfu recovered.
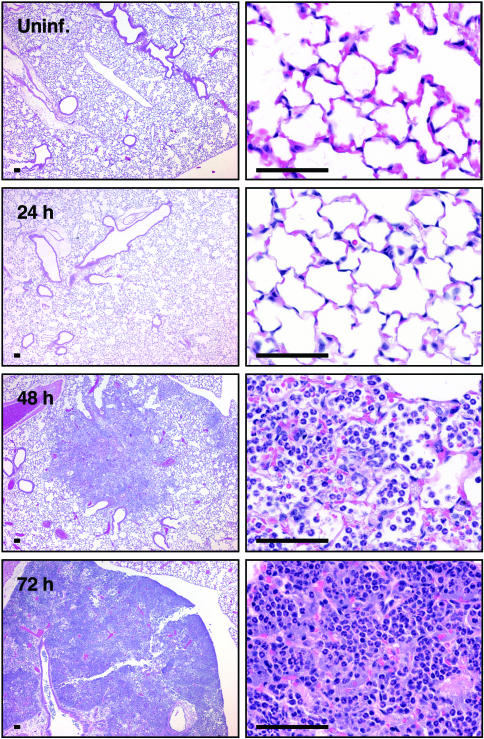
Histopathology of Mouse Lungs Infected with Y. pestis. To compare the effects of primary pneumonic plague on experimentally infected mice with that observed in humans, we examined the changes in pathology of the lungs of C57BL/6 mice at various times after infection with Y. pestis (Fig. 2). After 24 h, lungs looked unremarkable compared with uninfected tissue. We observed normal amounts of alveolar macrophages, although some appeared slightly activated as foamy macrophages with increased cytoplasm. By 48 h, however, changes in the lung architecture and types of cells present were visible compared with uninfected lungs. Although the larger bronchi remained relatively unaffected, the smaller bronchi appeared hyperplastic, with exudates present in the bronchi spreading outward into the alveoli. Foci of neutrophils loosely packing the alveoli were observed surrounding some of the smaller bronchi with a corresponding decrease in the number of alveolar macrophages compared with 24 h. Some slight hemorrhage was also evident, whereas other sections of lungs remained unremarkable, lacking any visible damage or inflammatory effects. At 72 h postinoculation, the mice had developed a purulent multifocal severe exudative bronchopneumonia. Entire lobes of the lungs appeared consolidated and had lost most of their recognizable architecture, although some of the larger bronchi were still visible. Along with extensive hemorrhage, large numbers of neutrophils were tightly packed within what remained of the alveolar structure.
Fig. 2.
Histology of lungs after intranasal infection with Y. pestis strain CO92. Lungs were inflated and fixed with 10% formalin, and 5-μm sections were stained with hematoxylin/eosin. Lungs were examined from uninfected mice or mice infected with 1 × 104 cfu Y. pestis for 24, 48, or 72 h. (Scale bars, 50 μm.)
Localization of Y. pestis Within Infected Mouse Lungs. In lung sections from individuals who died of primary pneumonic plague, Y. pestis is found mostly in the small airways and alveoli as extracellular bacilli (16, 17). To determine the localization of Y. pestis after the intranasal infection of mice, we used the Steiner silver stain to visualize bacteria in the lungs at various times during the infection. Consistent with the absence of pathological changes in the lungs at 24 h, we were unable to observe bacteria in the tissue at this point during the infection (Fig. 5, which is published as supporting information on the PNAS web site). By 48 h, however, bacteria were clearly visible within the alveoli and small bronchioles, primarily centered within and around the neutrophilic infiltrate. Overall, the bacteria were present as extracellular individual bacilli, and in some cases rings of bacteria appeared to encircle blood vessels. We did not observe bacteria within the interstitium of the alveoli. At 72 h after infection, large areas of the lungs were filled with masses of darkly staining material, in which individual bacilli could not be distinguished. These are likely large microcolonies of bacteria, although the presence of bacterial debris could not be excluded. In some sections, individual extracellular bacteria were visible. Although the larger bronchi appeared hyperplastic, they remained relatively free of bacteria.
Cytokine Production During Experimental Pneumonic Plague. The production of pro- and antiinflammatory cytokines by immunomodulatory cells is a crucial component of the host response to control bacterial infection. The pCD1-based type III secretion system (TTSS) of Y. pestis, however, is thought to subvert the immune response in part by altering the types and levels of cytokines produced during plague infection. A survey of the inflammatory cytokines produced in the lungs in response to Y. pestis, particularly during primary pneumonic plague, has not been reported. Therefore, we measured the levels of the cytokines IL-12p70, TNF, IFN-γ, monocyte chemoattractant protein (MCP-1), IL-10, and IL-6 in lung homogenates of mice at various times during infection with Y. pestis (Table 1). After 24 h, modest levels of most cytokines tested were detected, although in the majority of cases the amounts were not significantly different from uninfected mice. The levels of most cytokines rose significantly by 48 h, however, and continued to increase by day 3, particularly those of MCP-1 and IL-6. Of note, IL-10 remained undetectable throughout the infection with the exception of one mouse. To confirm the lack of IL-10 production during infection, we compared the mRNA levels of this cytokine in RNA extracted from infected lung tissue at 24 and 48 h with that present in uninfected lungs by qRT-PCR. We did not detect any significant induction of IL-10 by this method (data not shown).
Table 1. Cytokine levels in lung homogenates.
| Mean cytokine level (pg/g of tissue) ± SEM at
|
||||
|---|---|---|---|---|
| Cytokine | 0 h | 24 h | 48 h | 72 h |
| IL-12p70 | 52 ± 5 | 47 ± 5 | 137 ± 34 | 170 ± 23 |
| TNF | 31 ± 3 | 46 ± 7 | 1031 ± 251 | 1474 ± 174 |
| IFN-γ | ND | 57 ± 30 | 3715 ± 977 | 2871 ± 462 |
| MCP-1 | 246 ± 28 | 128 ± 15 | 1099 ± 350 | 22,056 ± 10,939 |
| IL-10 | ND | ND | ND | 1 ± 1 |
| IL-6 | 40 ± 11 | 505 ± 293 | 21,484 ± 5786 | 126,132 ± 7015 |
ND, not detected.
In Vivo Transcriptional Profile of Y. pestis During Primary Pneumonic Plague. The transmission of Y. pestis by an arthropod vector is unique among the Enterobacteriaceae. As such, adaptation of the bacterium from a flea to a mammalian host includes responses to changes in temperature (26-37°C), calcium, iron, and other factors. The transitions encountered in primary pneumonic plague are quite different, because this represents a mammal-to-mammal respiratory transition; the organisms are already adapted to 37°C, and the first cells they encounter are those lining the airway. Thus, an analysis of the global transcriptional responses of Y. pestis to a pulmonary environment is an important step toward understanding how the bacterium causes pneumonic plague. To this end, we used DNA microarray technology to profile the transcriptome of Y. pestis during experimental primary pneumonic plague using the intranasal mouse model of infection.
We designed oligonucleotide-based arrays representing 100% of the predicted ORFs of Y. pestis strain CO92 (18) and included control oligonucleotides for ORFs from selected mammals, viruses, and other bacterial pathogens. Unfortunately, the contamination of bacterial samples with eukaryotic RNA during host-pathogen interactions, particularly in vivo, can interfere with microarray hybridization and lead to false-positive signals (9). Therefore, because of the observation that Y. pestis infection of lung tissue is primarily extracellular, we compared the recovery of bacilli from infected mouse lungs by bronchoalveolar lavage (BAL) with that from homogenized lungs. Forty-eight hours after inoculation, lungs were either homogenized or lavaged with 3 × 1 ml of PBS, and cfu were determined. There was no significant difference in the numbers of bacteria recovered by either method (Fig. 6A, which is published as supporting information on the PNAS web site). RNA extracted from the BAL method, however, was vastly reduced in the levels of eukaryotic rRNA compared with those present from homogenized samples (ratio of 1:5 bacterial to eukaryotic rRNA by homogenization, 10:1 bacterial to eukaryotic rRNA by BAL) (Fig. 6B). Therefore, RNA extracted from BAL fluid 48 h postinfection was used for the microarray analysis.
RNA was isolated from BAL fluid recovered from two independent replicates of intranasal infections of 10 C57BL/6 mice each. Two sets of control RNA for array cohybridizations were extracted from bacteria grown for 12 h at 37°C in BHI broth containing 2.5 mM CaCl2, similar to conditions used to prepare Y. pestis for the animal inoculations. To increase the total amount of material available for hybridizations and downstream analyses, RNA was linearly amplified (Supporting Text). This procedure yielded, on average, ≈50-75 μg of antisense RNA from 0.5 μg of input RNA.
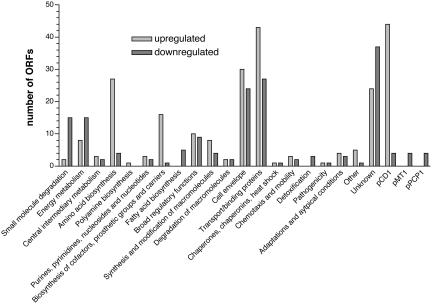
Of the 4,037 Y. pestis ORFs represented by oligonucleotides on the DNA microarray, 405, or 10% of the genome, were differentially regulated in the lungs of mice at 48 h (Table 2). Of these, 349 ORFs are encoded on the chromosome, whereas the remaining 56 are located on the three plasmids carried by Y. pestis strain CO92 (48 on pCD1; 4 on pMT1; 4 on pPCP1) (Fig. 3). Two hundred thirty-four ORFs were up-regulated, and 171 ORFs were downregulated in vivo compared with bacteria grown in BHI broth at 37°C. Based on the functional classification of genes annotated by the Sanger Institute (www.sanger.ac.uk/Projects/Y_pestis), the greatest number of transcriptional changes occurred in genes associated with the cell envelope (13% of the total; 30 up-regulated, 24 down-regulated), genes encoding transport and/or binding proteins (17% of the total; 42 up-regulated, 27 down-regulated), and those with unknown function (15% of the total, 24 upregulated, 37 down-regulated) (Table 3, which is published as supporting information on the PNAS web site). Additionally, 27 genes involved in amino acid biosynthesis were up-regulated, whereas only four were down-regulated in vivo. Among those up-regulated were genes associated with the histidine, pyruvate, glutamate, and aspartate families of amino acid biosynthesis. Also of note is the down-regulation in vivo of genes involved with small molecule degradation and energy metabolism, including those associated with the tricarboxylic acid cycle and ATP-proton motive force.
Fig. 3.
Functional classification of differentially regulated Y. pestis genes during primary pneumonic plague. Genes differentially regulated (48-h lungs vs. 37°C BHI broth) 2-fold or more in both RNA amplifications were divided into 20 categories based on the CO92 chromosomal annotation; genes differentially regulated on the three plasmids are indicated separately.
Multiple virulence-associated loci were also differentially regulated during the infection. Of the 234 Y. pestis ORFs up-regulated in vivo, 33 genes, or 14% of the total, are associated with the pCD1-based TTSS, essential to the virulence of Y. pestis. Both the iron acquisition system yersiniabactin (ybtETPQXS, irp1, irp2, and psn) and the hemin uptake operon (hmsHFRS), encoded by genes contained within the 102-kb pgm locus necessary for virulence via peripheral routes of infection (14), were up-regulated in the lungs of infected mice. Interestingly, the hms operon is thought to be posttranscriptionally regulated (19), although there may be environmental signals that affect the transcription of this system in vivo. Other virulence determinants, including the psa operon encoding the pH 6.0 antigen fimbrial structure (psaABEF) and the plasminogen activator protease Pla, were down-regulated in vivo at 48 h compared with broth-grown bacteria. The pH 6.0 antigen is thought to be expressed during intracellular association with macrophages (20) and is transcriptionally up-regulated in vitro at 37°C under low pH, whereas Pla is essential for the dissemination of the plague bacillus from peripheral sites of infection but unnecessary for virulence via the i.v. route (21). The contributions of Pla and the psa operon to pneumonic plague are unknown or unclear (22, 23), and thus further work to elucidate their roles during primary pulmonary infection is warranted.
Genes involved in the detoxification of reactive oxygen species, including katA, katY, and sodB, were also down-regulated in the lungs, suggesting that Y. pestis may have reduced exposure to these molecules at this point during the infection. Indeed, multiple genes involved in the stress response were differentially regulated in vivo, including the cold-shock responsive genes cspA1 and cspA2, both homologues of cspB (YPO1398 and YPO2659), cspD, the global stress responsive gene gsrA/htrA, and the flavohemoprotein gene hmp. As expected, the F1 capsule, encoded by the caf operon located on the pMT1 plasmid, was not differentially regulated in the mouse lung, because capsule production is induced at 37°C, the temperature at which Y. pestis was cultured in vitro for control RNA extraction.
Confirmation of Microarray Data by qRT-PCR. The validity of the microarray results was assessed by qRT-PCR. The fold expression change of 27 randomly selected, differentially regulated genes was determined from the same antisense RNA samples used for microarray hybridization. Overall, the change in expression of 89% of the genes tested by qRT-PCR was in agreement with the direction of fold change as determined by microarray analysis (Fig. 7, which is published as supporting information on the PNAS web site).
Discussion
Although the incidence of plague has decreased substantially over the last century, that Y. pestis infects humans via the inhalation of aerosolized particles and causes the rapid and devastating disease pneumonic plague has recently raised concern over the possible use of the bacterium as a bioterror agent. In comparison with the more common bubonic form of the disease, however, the molecular and cellular mechanisms that are responsible for primary pneumonic plague have been relatively understudied. Thus, the work presented here provides information on the progression of primary pneumonic plague caused by a fully sequenced strain of the Orientalis biovar in an inbred mouse model of infection using current technologies.
The pulmonary disease we observed in the C57BL/6 mouse strain is remarkably similar to that seen in humans in both progression and severity. In the first 24 h after infection, mice remain asymptomatic, although the bacterial load in the lungs increased ≈10-fold every 12 h. Interestingly, early studies of pneumonic plague in mice showed a reproducible decrease in cfu in the lungs until ≈24 h after inoculation (8, 24). This discrepancy with our results may be caused by multiple factors, including differences in the route of infection (aerosol vs. intranasal), strain of Y. pestis (139L or EV-76 vs. CO92), strain of mouse (male Namru albino vs. female C57BL/6), or bacterial growth condition for preparation of the inoculum (room temperature vs. 37°C). Indeed, the temperature-dependent up-regulation of multiple antiphagocytic factors at 37°C may be the most likely explanation for these differences in the kinetics of infection, because the bacteria prepared under our conditions more closely mimics those in the mammal-to-mammal transmission during primary pneumonic plague (37-37°C) and are therefore primed to prevent phagocytosis.
The asymptomatic state of the infection in the first 24 h corresponds with the absence of histological changes in the lungs or dramatic alterations in inflammatory cytokine levels. The relative quiescence at this point is likely because of the potent antiinflammatory activity of the pCD1-based TTSS, particularly against alveolar macrophages. Indeed, Nakajima and Brubaker (25) observed robust TNF and IFN-γ production in the spleens of pCD1- Y. pestis-infected mice early in the infection but not when mice were infected with pCD1+ bacteria. The relatively high intranasal LD50 compared with the LD50 via the s.c. route (one bacillus), however, suggests that significant impediments within the respiratory tract, whether immune response, mechanical interference, and/or clearance (8), or other factors can affect the outcome of the disease, and that the mere introduction of the bacteria into the airway is not sufficient to establish a lethal infection.
Although primary pneumonic plague results in pulmonary collapse, dissemination of Y. pestis from the lungs to other tissues and organs may also influence the progression of the disease. Beginning 36 h after intranasal inoculation, we detected viable bacteria in the spleens of infected mice, indicating that Y. pestis had escaped the confines of the respiratory system. Interestingly, this correlated with a 3-log increase in Y. pestis cfu in the lungs between 24 and 36 h postinfection. The presence of bacteria surrounding blood vessels within the lungs at 48 h suggests that the dramatic rise in cfu may be caused by the recirculation of bloodborne Y. pestis back into the lungs, creating additional foci of infection within previously uncolonized areas. Thus, what we characterize as primary pneumonic plague at this stage of the disease may in fact be a combination of primary and secondary foci of infection.
By 48 h, the development of visible symptoms of pneumonic plague corresponded with a shift to a more inflammatory state in the lungs, in contrast to the relatively pre- or antiinflammatory condition just 1 day earlier. Numerous extracellular bacilli in the alveoli and small airways were often concomitant with an inflammatory infiltrate consisting primarily of neutrophils. Likewise, we measured a significant rise in multiple proinflammatory cytokines and chemokines at this time, including both TNF and IFN-γ, suggesting a vigorous (yet ultimately unsuccessful) host response to the infection. Although we detected few intracellular bacteria within neutrophils, the migration and activation of these immune cells, including the release of reactive nitrogen species and neutrophil elastase, may cause considerable damage to the host and may be at least partially responsible for the deterioration in the condition of the animals. By 72 h, the consolidation of entire lobes of the lungs and the resulting edema is likely to cause a significant impediment in oxygen exchange.
The development of a proinflammatory state within the lungs of Y. pestis-infected mice prompted us to begin studies to explore the adaptation of the bacterium to this potentially hostile environment. As an initial step toward this long-term goal, we chose to analyze the transcriptional profile of the plague bacillus in the lungs of infected mice by using DNA microarray technology. Two significant obstacles to using DNA-based microarrays for the analysis of bacterial transcription in vivo are the potential for excess amounts of eukaryotic RNA to be copurified during the isolation procedure that might contribute to nonspecific hybridization on the arrays and the relatively low levels of bacterial RNA isolated from host tissue, particularly early in the infection. We addressed the first problem by isolating bacteria from the mouse lung by BAL, which was essentially equivalent to homogenization of the tissue in terms of bacterial recovery yet reduced the contamination of eukaryotic RNA to <10% of the total RNA purified. It is important to note, however, that although similar numbers of bacteria were recovered from the lungs by the two methods, there may be a subset of Y. pestis that remains in the lungs and is unrecoverable by BAL, thereby altering the total transcriptional profile of the recovered bacteria slightly. The second obstacle was overcome by in vitro transcription (IVT)-mediated linear amplification of the purified RNA, a commonly used technique for the amplification of eukaryotic RNA (26). This was performed by polyadenylating the purified RNA with the enzyme poly(A) polymerase and then reverse-transcribing the population with oligo(dT) primers containing a T7 RNA polymerase promoter. Thus, the RNA extracted from Y. pestis isolated by BAL, combined with IVT-mediated linear amplification, produced sufficient RNA from 10 mice per biological replicate for multiple array hybridizations. In addition, random assessment of the differential expression of 27 genes by qRT-PCR showed 89% concordance with the microarray data, validating the use of BAL in this system to recover bacterial RNA relatively free of eukaryotic RNA that might nonspecifically hybridize with the arrays.
The remarkable capacity of Y. pestis to replicate essentially unchecked by the host immune response is attributed to the antiphagocytic and antiinflammatory effects of the TTSS encoded on pCD1. This system is expressed in vitro at 37°C in the absence of calcium, and effector molecules are released into the culture medium (for review, see ref. 27). At 37°C in the presence of calcium, the system is “primed” in that the yop-ysc regulon is up-regulated over 26°C, but secretion is prevented (28, 29). Indeed, a comparison of the transcriptional response of Y. pestis grown in BHI medium containing 2.5 mM CaCl2 at 37°C showed the up-regulation of many TTSS genes compared with 26°C (unpublished data). Our DNA microarray analysis of bacteria grown in vivo, however, revealed additional up-regulation of the pCD1-based TTSS when compared with in vitro growth at 37°C. This indicates that there may be signals in vivo that further induce the system, the most likely being host cell contact, that stimulate a positive feedback mechanism (30). Interestingly, lcrF, a transcriptional activator that controls expression of the TTSS (31), was down-regulated 3-fold, suggesting that the modulation of the system in vivo may be more sophisticated than previously appreciated. Nonetheless, the observation that a majority of these genes are up-regulated during pneumonic plague as determined by DNA microarray analysis provides biologic validation of the system. In addition, these data suggest that the TTSS may delay the host inflammatory response and later contribute to the survival of the bacteria in a highly proinflammatory environment.
Conclusion
Y. pestis causes a purulent multifocal severe exudative bronchopneumonia in mice when administered via the intranasal route. Much like in humans, the disease is rapid and severe, resulting in death of the animal within 3-4 days. Although bacterial proliferation is rapid, the initial stage of the infection is relatively quiescent and antiinflammatory in nature. This develops rapidly into a highly proinflammatory state as cytokine and chemokine levels rise, dissemination of the bacterium to the spleen occurs, and mice become symptomatic. Also, the use of DNA microarray technology to measure the transcriptional profile of Y. pestis during infection has provided a valuable tool to assess the global response of the bacterium during the disease. Thus, this model system should be useful for the continued study of primary pneumonic plague as a distinct disease of mammals caused by Y. pestis, providing perspectives from both the host and the bacterium.
Supplementary Material
Acknowledgments
We thank Drs. Craig Rubens and Marie LaRegina for their helpful comments, Michael Heinz and Christopher Sawyer of the Washington University Genome Sequencing Center Microarray Core for the microarray hybridizations, and the Washington University Digestive Diseases Research Core (DDRCC) Facility for the histological sections. This work was funded by National Institutes of Health (NIH) Grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) and by NIH Grant AI53298. The DDRCC core is supported by NIH Grant DK52574. W.W.L. was supported by the Infectious Diseases Training Grant to Washington University and by the Clinical/Translational Fellowship Program of the MRCE.
Author contributions: W.W.L., V.L.M., and W.E.G. designed research; W.W.L. and S.D.C. performed research; W.W.L. contributed new reagents/analytic tools; W.W.L., S.D.C., V.L.M., and W.E.G. analyzed data; and W.W.L. and W.E.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BHI, brain-heart infusion; cfu, colony-forming units; qRT-PCR, quantitative RT-PCR; BAL, broncho-alveolar lavage; TTSS, type III secretion system.
References
- 1.Inglesby, T. V., Dennis, D. T., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Friedlander, A. M., Hauer, J., Koerner, J. F., et al. (2000) J. Am. Med. Assoc. 283. [DOI] [PubMed]
- 2.Guarner, J., Shieh, W.-J., Greer, P. W., Gabastou, J.-M., Chu, M., Hayes, E., Nolte, K. B. & Zaki, S. R. (2002) Am. J. Clin. Pathol. 117, 205-209. [DOI] [PubMed] [Google Scholar]
- 3.Doll, J. M., Zeitz, P. S., Ettestad, P., Bucholtz, A. L., Davis, T. & Gage, K. (1994) Am. J. Trop. Med. Hyg. 51, 109-114. [DOI] [PubMed] [Google Scholar]
- 4.Werner, S. B., Weidmer, C. E., Nelson, B. C., Nygaard, G. S., Goethals, R. M. & Poland, J. D. (1984) J. Am. Med. Assoc. 251, 929-931. [PubMed] [Google Scholar]
- 5.Finegold, M. J. (1969) Am. J. Pathol. 54, 167-185. [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer, K. F., Quan, S. F. & Larson, A. (1948) Am. Rev. Tuberc. Resp. Dis. 57, 312-321. [DOI] [PubMed] [Google Scholar]
- 7.Smith, P. N. (1959) J. Infect. Dis. 104, 78-84. [DOI] [PubMed] [Google Scholar]
- 8.Smith, P. N., McCamish, J., Seely, J. & Cooke, G. M. (1957) J. Infect. Dis. 100, 215-222. [DOI] [PubMed] [Google Scholar]
- 9.Mangan, J. A., Monahan, I. M. & Butcher, P. D. (2002) in Functional Microbial Genomics, eds. Wren, B. & Dorrell, N. (Academic, Amsterdam), Vol. 33, pp. 137-151. [Google Scholar]
- 10.Snyder, J. A., Haugen, B. J., Buckles, E. L., Lockatell, C. V., Johnson, D. E., Donnenberg, M. S., Welch, R. A. & Mobley, H. L. (2004) Infect. Immun. 72, 6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talaat, A. M., Lyons, R., Howard, S. T. & Johnston, S. A. (2004) Proc. Natl. Acad. Sci. USA 101, 4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revel, A. T., Talaat, A. M. & Norgard, M. V. (2002) Proc. Natl. Acad. Sci. USA 99, 1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, S., Bearden, S. W., Geoffroy, V. A., Fetherston, J. D. & Perry, R. D. (2001) Infect. Immun. 69, 2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry, R. D. & Fetherston, J. D. (1997) Clin. Microbiol. Rev. 10, 35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer, K. F. (1950) J. Immunol. 64, 139-163. [PubMed] [Google Scholar]
- 16.Dennis, D. T. & Meier, F. T. (1997) in Pathology of Emerging Infections, eds. Horsburgh, C. R. & Nelson, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 21-47.
- 17.Strong, R. P., Crowell, B. C. & Teague, O. (1912) Philipp. J. Sci. B. Philipp. J. Trop. Med. 7, 203-221. [Google Scholar]
- 18.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T. G., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., Baker, S., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 19.Perry, R. D., Bobrov, A. G., Kirillina, O., Jones, H. A., Pedersen, L., Abney, J. & Fetherston, J. D. (2004) J. Bacteriol. 186, 1638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindler, L. E. & Tall, B. D. (1993) Mol. Microbiol. 8, 311-324. [DOI] [PubMed] [Google Scholar]
- 21.Sodeinde, O. A., Subrahmanyam, Y. V., Stark, K., Quan, T., Bao, Y. & Goguen, J. D. (1992) Science 258, 1004-1007. [DOI] [PubMed] [Google Scholar]
- 22.Welkos, S., Pitt, M. L. M., Martinez, M., Friedlander, A., Vogel, P. & Tammariello, R. (2002) Vaccine 20, 2206-2214. [DOI] [PubMed] [Google Scholar]
- 23.Worsham, P. L. & Roy, C. (2003) Adv. Exp. Med. Biol. 529, 129-131. [DOI] [PubMed] [Google Scholar]
- 24.Smith, P. N. (1959) J. Infect. Dis. 104, 85-91. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima, R. & Brubaker, R. R. (1993) Infect. Immun. 61, 23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D. & Eberwine, J. H. (1990) Proc. Natl. Acad. Sci. USA 87, 1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornelis, G. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goguen, J. D., Walker, W. S., Hatch, T. P. & Yother, J. (1986) Infect. Immun. 51, 788-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straley, S. C. & Perry, R. D. (1995) Trends Microbiol. 3, 310-317. [DOI] [PubMed] [Google Scholar]
- 30.Pettersson, J., Nordfelth, R., Dubinina, E., Bergman, T., Gustafsson, M., Magnusson, K. E. & Wolf-Watz, H. (1996) Science 273, 1231-1233. [DOI] [PubMed] [Google Scholar]
- 31.Lambert de Rouvroit, C., Sluiters, C. & Cornelis, G. R. (1992) Mol. Microbiol. 6, 395-409. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.