Abstract
During the transition from an initiation complex to an elongation complex (EC), the single-subunit bacteriophage T7 RNA polymerase (RNAP) undergoes dramatic conformational changes. To explore the significance of these changes, we constructed mutant RNAPs that are able to form disulfide bonds that limit the mobility of elements that are involved in the transition (or its reversal) and examined the effects of the crosslinks on initiation and termination. A crosslink that is specific to the initiation complex conformation blocks transcription at 5–6 nt, presumably by preventing isomerization to an EC. A crosslink that is specific to the EC conformation has relatively little effect on elongation or on termination at a class I terminator (Tφ), which involves the formation of a stable stem–loop structure in the RNA. Crosslinked ECs also pause and resume transcription normally at a class II pause site (concatamer junction) but are deficient in termination at a class II terminator (PTH, which is found in human preparathyroid hormone gene), both of which involve a specific recognition sequence. The crosslinked amino acids in the EC lie close to the upstream end of the RNA–DNA hybrid and may prevent a movement of the polymerase that would assist in displacing or releasing RNA from a relatively unstable DNA–RNA hybrid in the paused PTH complex.
Keywords: crosslink, transcription
Although it consists of a single subunit, T7 RNA polymerase (RNAP) carries out all of the steps in the transcription cycle in the same manner as multisubunit RNAPs (1). Therefore, it provides a useful model for studies of the fundamental aspects of transcription. As is the situation with other RNAPs, T7 RNAP forms an unstable initiation complex (IC) that synthesizes and releases short abortive transcripts before it isomerizes to a stable elongation complex (EC) (2). The transition to an EC is accompanied by major structural rearrangements in the protein, resulting in the formation of elements that are important for the stability and processivity of the EC. These include a cavity that can accommodate an RNA–DNA hybrid of 8–9 bp and pores for RNA exit and substrate entry (3, 4). These features of the T7 RNAP EC are similar to those of the multisubunit RNAPs (5, 6).
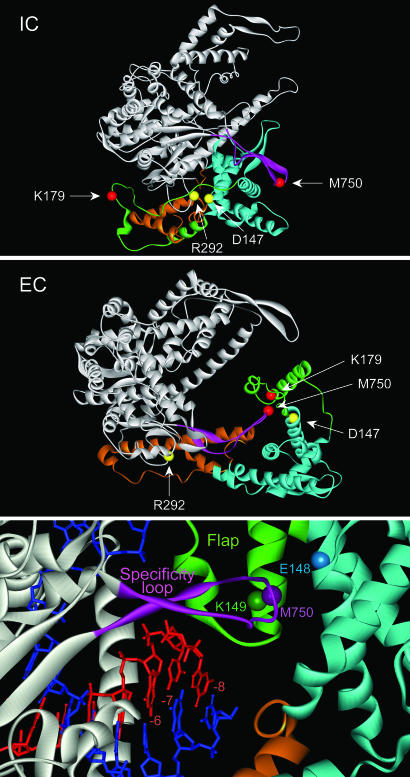
The structural rearrangements leading to an EC largely involve the N-terminal domain of the RNAP (3, 4). In particular, a “core” subdomain (residues 72–151 and 206–257) rotates and translocates 35 Å as a rigid body to allow room for the RNA–DNA hybrid. Other regions in the N-terminal domain become refolded to form an element that interacts with the RNA–DNA hybrid and participates in RNA displacement and resolution of the transcription bubble (the “flap” subdomain, residues 152–205). A structural element in the C-terminal domain (the specificity loop, residues 739–769) also becomes rearranged during the transition. Whereas in the IC this element is involved in promoter contacts, in the EC it becomes associated with the displaced transcript and forms part of the RNA exit pore (7, 8) (see Fig. 1).
Fig. 1.
Structures of T7 RNAP IC and EC. Structures of the RNAP in the IC (Top) and EC (Middle) conformations (Protein Data Bank ID 1qln and 1h38) are presented as ribbons and colored as follows: C-terminal domain (residues 267–883), gray; flap subdomain (152–205), green; specificity loop (739–769), pink; core subdomain (72–151, 206–257), blue; and other elements of the N-domain, brown. The α-carbons of key residues are represented as colored spheres. The orientation of both structures with regard to the unrearranged C-terminal domain is the same. (Bottom) A close-up view of the RNA exit pore is shown; DNA is blue, RNA is red, positions of nucleotides in the RNA–DNA hybrid are indicated relative to the 3′ end of the RNA (at –1).
T7 RNAP pauses or terminates at two types of signals. Class I termination signals, such as the Tφ terminator located in the late region of the T7 genome, encode an RNA with a stable stem–loop structure followed by a run of U residues. This type of signal is similar to the intrinsic terminators used by bacterial RNAPs, and indeed, T7 RNAP has been found to use a number of such signals, suggesting a common mechanism of termination (9–11). Class II pause/termination signals such as PTH, the termination site for T7 RNAP found in human preparathyroid hormone gene, do not involve an apparent RNA structure, but require a conserved 8-bp sequence [(C/U)AUCUGU(U/A)] encoded in the DNA template (11–15). Although the class II signal located in the concatamer junction (CJ) of T7 DNA serves as a strong pause site that does not give rise to efficient termination, the presence of additional U residues downstream of the conserved sequence results in efficient termination (13, 14, 16). Termination at class I and class II signals appears to involve separate mechanisms, as mutations of the RNAP that prevent termination at class II signals do not affect termination at class I signals (9, 17, 18).
To explore the importance of conformational changes in T7 RNAP during transcription initiation and termination, we constructed mutant RNAPs that are able to form disulfide bonds that are specific to the IC or EC conformations and examined the effects of these crosslinks on transcription.
Materials and Methods
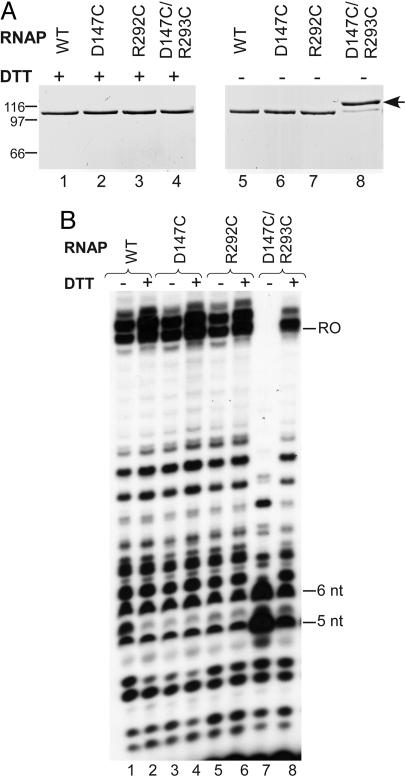
Design and Construction of Mutant T7 RNAPs. T7 RNAP structures (Protein Data Bank ID codes 1aro, 1cez, 1qln, 1h38, and 1msw) were examined with disulfide by design (19). The double mutants D147C/R292C and K179C/M750C were predicted to form Cys–Cys crosslinks in IC or EC conformations, respectively. Mutant RNAPs with Cys substitutions at each of these locations were constructed by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis kit, Stratagene). The mutant enzymes were purified as described (20) except that 2-mercaptoethanol was omitted during the final stage of purification (elution from a phosphocellulose column). Formation of disulfide bond linkage was monitored by electrophoresis in SDS/10% PAGE gels (Fig. 2) or 4–12% NuPAGE Bis-Tris gels (Invitrogen, Fig. 3) in the presence or absence of 5 mM DTT as indicated. Different electrophoresis conditions were needed to resolve the two different crosslinked proteins from protein without crosslinks, and the apparent fraction of crosslinked protein was somewhat variable.
Fig. 2.
Effects of disulfide linkages in the IC. (A) WT and mutant RNAPs purified under nonreducing conditions were analyzed by gel electrophoresis in the presence or absence of DTT; positions of Mr markers are indicated at the left. (B) A DNA template that contains a T7 promoter and produces a 20-nt runoff product (MJ6/MJ7) was transcribed by the RNAP indicated, and the products were resolved by gel electrophoresis. [T7 RNAP often adds a nontemplated base to the transcript, resulting in an additional runoff band of 21 nt (39).]
Fig. 3.
Effects of disulfide linkages in the EC on termination. (A) RNAPs indicated were incubated for 10 min in the absence of nucleic acid (free RNAP; lanes 1, 4, and 7), with a nucleic acid scaffold that allows the formation of an EC (4, 21) (lanes 2, 5, 8, 11, and 13) or with a promoter template (MK61/MK62) under conditions that allow formation of a binary complex (lane 9) or an EC halted at +22 (EC22) (lanes 3, 6, 10, 12, and 14), and the samples were analyzed by electrophoresis under nonreducing conditions (lanes 1–10). Samples in lanes 11–14 were exposed to 5 mM DTT either before (lanes 11 and 12) or after (lanes 13 and 14) assembly of the complex. (B) A DNA template that contains a class II (PTH) termination signal downstream from a T7 promoter was incubated with the RNAP indicated in the presence of GTP, [α-32P]ATP, and UTP for 1 min to allow the formation of a transcription complex halted at +23. Heparin (100 μg/ml) was added to prevent further initiation, and after 1 min the reaction was completed by addition of CTP to allow runoff transcription. RNA samples were analyzed by gel electrophoresis. The positions of the terminated (349 nt) and runoff (541 nt) products are indicated in the margins. Reactions were carried out in the presence or absence of 5 mM DTT, as indicated. (C) Same as B, except the DNA template contained a class I termination signal (Tφ).
Template Preparation and Transcription Assays. A nucleic acid scaffold that allows the formation of an EC by T7 RNAP (TS1/NT1/RNA12) has been described (21). Synthetic double-stranded templates that contain a T7 promoter were constructed by annealing complementary T and NT strands together at 70°C for 10 min followed by slow cooling to room temperature (22). The sequences of the NT strands (5′–3′) for each pair of oligomers were as follows: MJ6/MJ7 (Fig. 2), TCGAAATTAATACGACTCACTATAGGGAGACCACAACCTCTCGT; MK61/MK62 (Fig. 3), ATCTGCAGTAATACGACTCACTATAGGGAGAGTAAGTAGGTAGTGTACCTACGTAGCTAACTGCGGTCC; NM33/NM34 (Fig. 4), CGCTAATACGACTCACTATAGGGAGAGAAGAGTGTAGTATATCTGTTACAGTCTCCTGGCTCAACCG; MK89/MK90 (Fig. 4), CGCTAATACGACTCACTATAGGGAGAGAAGAGTGTAGTATATCTGTTTTCTTGCAAGGGCTCAACCG. Templates that contain a class I or II termination signal downstream from a T7 promoter were generated by PCR amplification from plasmids pMK25 (PTH) or pMK26 (Tφ) by using MK56 (GCTTATCATCGATCTGCAGTA ATACGACTC) and MK57 (GGTGCACGGCCCACGTGGCCAC) as primers; the templates were purified by using a QIAquick nucleotide removal kit (Qiagen). pMK25 and pMK26 were derived from pDL42 and pDL44 (17) site-directed mutagenesis to provide a promoter with a downstream sequence that allows the formation of a halted complex at +23 (sequences available upon request). DNA and RNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and Dharmacon (Chicago), respectively.
Fig. 4.
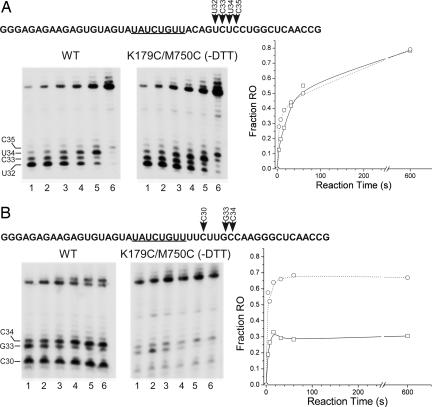
Effects of disulfide linkages in the EC on pausing and termination. (A) A template that contains the CJ class II pause signal (underlined) was assembled by annealing together the NT and T strand oligomers NM33 and NM44; the transcribed sequence downstream from the promoter is given at the top. The template (0.25 μM) was incubated with either WT RNAP in the presence of 5 mM DTT or with K179C/M750C in the absence of DTT (0.12 μM each RNAP) in the presence of GTP (300 μM), ATP (10 μM), and UTP (10 μM) for 2 min to allow formation of an EC halted at +22. The halted complexes were then chased by the addition of 10 μM CTP, 100 μg/ml heparin, and 1 μCi [α-32P]ATP. Samples were removed at 4, 8, 16, 32, 60, or 600 sec thereafter (lanes 1–6), mixed with stop buffer, and analyzed by electrophoresis. Positions of pause sites in the transcribed sequence (+32 to +35) are indicated. The fraction of products found at the runoff position (runoff/runoff + U32–C35) vs. time is plotted at the right (WT, □ and mutant RNAP, ○). (B) Same as A except that the template contains the PTH class II termination signal (oligos MK89 and MK90).
For the reactions shown in Fig. 2, 20 ng of T7 RNAP in transcription buffer (20 mM Tris·HCl, pH 7.9/10 mM MgCl2/0.1 mM EDTA/0.05% Tween 20) was incubated for 15 min at room temperature in the presence or absence of 5 mM DTT, as indicated. Transcription was initiated by the addition of 0.1 μM template (MJ6/MJ7), 0.5 mM NTPs, and 2 μCi (1 Ci = 37 GBq) [γ-32P]GTP to give a final volume of 10 μl. The reactions were terminated after 10 min by the addition of 10 μl of stop buffer [7 M urea/50 mM EDTA/0.02% (wt/vol) bromophenol blue/0.02% (wt/vol) xylene cyanol], and the products were resolved by electrophoresis in 20% (wt/vol) polyacrylamide gels containing 7 M urea. For termination assays (Fig. 3), elongation complexes halted at +23 (EC23) were formed by incubating 20 ng of enzyme, 0.1 μM PCR template, 0.3 mM GTP, 2 μCi [α-32P]ATP, and 0.1 mM ATP and UTP in the presence or absence of 5 mM DTT for 2 min. Heparin (100 μg/ml) was added to prevent further initiation, and after 1 min, CTP (0.1 mM) was added to drive completion of transcription. Transcription was stopped after 2 min, and the products were analyzed by electrophoresis in 6% (wt/vol) polyacrylamide gels containing 7 M urea.
Results
Design and Construction of Mutant RNAPs. A computer program (19) was used to identify amino acid pairs in the RNAP that are likely to allow the formation of disulfide bonds when substituted by Cys (Fig. 1). Two pairs that are characteristic of the IC or EC configurations were selected. Whereas residues D147 and R292 are close together in the IC (Cα distance of 5.4 Å), they become widely separated (Cα distance 43 Å) in the EC because of movement of the core subdomain. In contrast, residues K179 and M750 are widely separated in the IC (60 Å) but come close together in the EC (5.7 Å). The proximity of the latter two residues in the EC reflects the interaction of elements that are involved in the formation and maintenance of the RNA exit pore (the flap subdomain and the specificity loop, see Fig. 1) (3, 4, 7, 23).
Crosslinking the RNAP in the IC Conformation Blocks Transcription Beyond 5–6 nt. Mutant RNAPs involving residues D147 and R292 were constructed and purified in the normal fashion, except that reducing agent (2-mercaptoethanol) was omitted during the final stages of purification to allow formation of Cys–Cys bonds. Under nonreducing conditions the WT enzyme purified by this method migrates as a single band, as did each of the mutant enzymes involving a single Cys substitution (D147C and R292C; Fig. 2). In contrast, ≈80% of the mutant enzyme having both Cys substitutions (D147C/R292C) migrated more slowly than the WT enzyme under these conditions. Exposure of the D147C/R292C RNAP to the reducing agent DTT restored its mobility to that of the WT enzyme, consistent with the formation of a reversible intramolecular disulfide bond in this enzyme.
We next examined the transcription properties of the WT and mutant RNAPs in the presence or absence of DTT (Fig. 2B). Transcription by WT RNAP resulted in characteristic synthesis of abortive products 3–14 nt in length and a runoff product of 20 nt under both conditions, as did each of the single mutant RNAPs (lanes 1–6). In contrast, transcription by the crosslinked RNAP was strongly inhibited beyond a length of 5–6 nt (lane 7). This effect was largely eliminated when the enzyme was exposed to DTT (lane 8; the residual increase in synthesis of the 5- to 6-nt products is likely due to incomplete reduction of the crosslink under these conditions). We conclude that formation of the C147–C292 crosslink blocks the transition to an EC, commencing at 5–6 nt.
Effects of Crosslinking the RNAP in the EC Conformation. Structural data indicated that substitution of residues K179 and M750 with Cys should allow the formation of a disulfide bond when the enzyme is in the EC conformation, but not in the IC conformation (Fig. 1). To examine this, a mutant RNAP in which both of these residues were substituted with Cys (K179C/M750C) was incubated either in the absence of nucleic acids (free RNAP), under conditions that allow formation of a binary (RNAP/promoter) complex, or under conditions that lead to the formation of a stable EC (Fig. 3). Only those samples that would be expected to allow the formation of an EC resulted in the appearance of a protein band with altered mobility (Fig. 3A, lanes 8 and 10). Exposure of these samples to DTT either after the formation of the EC (Fig. 3A, lanes 13 and 14) or before EC formation (Fig. 3A, lanes 11 and 12) prevented the appearance of this band, consistent with the formation of a reversible crosslink that forms only in the EC. Substitution of both K179 and M750 was required to form the crosslink because neither of the single mutants (K179C or M750C) showed this effect (Fig. 3A, lanes 1–6).
To examine the consequences of the EC crosslink on transcription, we formed halted ECs on templates that contained either a class I (Tφ) or class II (PTH) termination signal downstream from a T7 promoter, allowed a brief time for formation of the crosslink, and then chased these complexes by adding the remaining NTPs (Fig. 3 B and C). The crosslinked complexes were able to resume transcription, as evidenced by the formation of runoff products. Moreover, all complexes terminated normally at the class I signal (Fig. 3C).
Strikingly, RNAPs crosslinked in the EC conformation were defective in termination at the class II signal (Fig. 3B, lane 7). Termination was restored by addition of DTT before the chase, indicating that it is the formation of the disulfide bond that is responsible for this effect (Fig. 3B, lane 8). Neither of the single mutants (K179C or M750C) was defective in class II termination (Fig. 3B, lanes 3–6), demonstrating that it is the formation of a disulfide bond between these two residues that is responsible for the termination defect.
A number of lines of evidence suggest that pausing and termination at class II signals is a multistage process in which binding of the conserved sequence followed by further transcript extension leads to pausing of transcription and accumulation of strain and possible conformational changes, which are then resolved either by resumption of transcription or by termination (15, 24, 25). The class II signal used in the experiments shown in Fig. 3 contains the conserved recognition sequence followed by a U-rich region (Fig. 4B), a configuration that was first identified in a cloned copy of the human preparathyroid hormone gene (26). The downstream U-rich region appears to favor termination, and most transcription complexes that encounter the PTH signal rapidly terminate (14). By contrast, the class II signal found in the concatemer junction (CJ) of replicating T7 DNA (Fig. 4A) lacks an extended U-rich region, and most transcription complexes that encounter this signal pause and then slowly resume transcription (14, 16). The different responses of ECs at these two signals is likely to reflect differences in stability of the paused complexes caused by the U-rich region, which is known to decrease the stability of an RNA–DNA hybrid (27).
To examine further the effects of crosslinking of the EC on pausing and termination, we determined the kinetics of transcription on synthetic templates that contained either a CJ or PTH signal. Startup complexes halted at +22 (EC22), formed as in Fig. 3, were chased by addition of the remaining NTPs, and samples were withdrawn at different times (Fig. 4).
WT and crosslinked (K179C/M750C) ECs had similar kinetics of pausing and resumption of transcription at the CJ site, i.e., most complexes paused at the CJ site at 4 s, few paused complexes released their RNA, and resumption of transcription proceeded slowly and continued well past 60 s (Fig. 4A). Apparently, recognition of the class II sequence, pausing, and resumption of transcription at the CJ site are barely, if at all, impaired by the crosslink.
Consistent with previous results (14), termination by WT ECs at the PTH site was rapid, and ≈70% of the complexes terminated within 16 sec (Fig. 4B). Nevertheless, synthesis of runoff transcripts increased between 4 and 16 sec, indicating that a significant fraction of ECs paused at the PTH site resumed transcription, although at a much faster rate than escape from the CJ pause. A smaller population of complexes paused at G33 appeared to resume transcription over a longer time period (similar to that observed at CJ). These features appear to be characteristic of pausing and termination at the class II PTH signal: rapid termination and/or escape from the signal for most complexes and slower escape for a minority of complexes. Thus, it appears that the extended U-run downstream of the signal results in both increased termination efficiency as well as a reduction in the length of the pause for most complexes.
The behavior of the crosslinked ECs was similar to that of the WT enzyme except that only ≈30% of complexes terminated at the PTH signal (as opposed to ≈70% for the WT enzyme). Again, termination was rapid, and most of the complexes that did not terminate resumed transcription after 4 sec.
The observation that the kinetics of pausing were nearly identical for the WT and crosslinked enzymes at both the CJ and termination signals indicates that the crosslink does not interfere with pausing, but with termination. As noted below, the crosslink involves elements in the flap domain and the specificity loop whose interactions are critical to the formation of the RNA exit pore and RNA displacement. Presumably, altering the mobility or flexibility of these elements impairs transcript displacement and/or release by complexes that are paused in an unstable environment provided by the U-rich region.
Discussion
For many enzymes, conformational changes may accompany important transition states, and the ability to perturb or restrain these transitions by using reversible disulfide bonds offers a potentially powerful method to examine the importance of these changes. Two quite distinct conformations for T7 RNAP have been reported. In free RNAP, and during the early stages of transcription (promoter binding and synthesis of transcripts <3 nt), the conformation of the enzyme remains largely unchanged (the IC conformation) (28, 29). The EC structure appears to be greatly stabilized by interactions of the protein with downstream DNA, the RNA–DNA hybrid, and the emerging transcript (21), and is observed only when the enzyme is in a context that provides these elements (3, 4). Consistent with these results, the formation of a disulfide bond that is characteristic of the IC (D147C–R292C) was observed in the absence of any nucleic acids, but the formation of a disulfide bond that is characteristic of the EC (K179C–M750C) was observed only in a halted EC or in the presence of a nucleic acid scaffold that supports the EC conformation (Figs. 2 and 3).
Although structural information reveals the configuration of the complexes before and after the transition, little is known about the pathway of the reorganization or the timing of these events. Two models have been proposed to describe the transition to an EC. One suggests that the length of the RNA–DNA hybrid remains fixed at ≈3 bp and/or that local conformational changes in the RNAP and “scrunching” of the template allow extension of the transcript up to 8–9 nt, whereupon a major reorganization of the protein occurs (3, 30). The other model proposes a gradual refolding of the RNAP to accommodate a growing RNA–DNA hybrid as it is extended from 3 bp (as observed in the IC) to 8–9 bp (as observed in the EC) (3, 4, 31). Support for a model in which the length of the RNA–DNA hybrid increases incrementally during the IC to EC transition comes from fluorescence assays, which indicate that the 5′ end of the growing RNA chain remains in the heteroduplex up to a chain length of 10 (32).
The 3-bp RNA–DNA hybrid observed in the IC structure is in the posttranslocated state (in which the 3′ end of the transcript has been displaced from the active site). This structure can accommodate the incorporation of an additional nucleotide (resulting in a hybrid length of 4 bp) without significant steric clash. However, translocation of the active site and subsequent extension of the hybrid could not occur without movement of the core subdomain, so it had earlier been suggested that relocation of the core subdomain would be required to accommodate growth of the transcript beyond 4 nt (4, 31). The observation that the D147C–R292C crosslink blocks extension of the RNA chain past 5–6 nt is consistent with the idea that movement of the core domain is necessary to accommodate extension of the RNA chain, as the crosslink prevents movement of residue 147 (in the core subdomain) away from residue 292 (in the C-terminal domain). In the natural IC, residues D147 and R292 form a salt bridge, but this interaction appears not to be critical for transcription, as the D147C and R292C substitutions (this work) and a D147A mutation (33) exhibit a normal pattern of transcription. We earlier observed that the mutation E148A, adjacent to D147 in the core subdomain, also interferes with extension of the RNA chain past 5 nt (33). E148 forms a salt bridge with another residue in the core, R155, an interaction that may help to stabilize the core conformation during the IC to EC transition. After conversion to an EC, residues 147 and 148 lie near the tip of the rearranged specificity loop and may help to stabilize the RNA exit pore (Fig. 1). The similar effects of the D147C–R292C crosslink and the E148A mutation suggest that rearrangement to form the RNA exit pore might normally be underway by the time the RNA chain reaches 5–6 nt in length.
The end of each cycle of transcription is punctuated by termination, in which the RNAP releases the transcript and dissociates from the template. At some point, the enzyme must revert from the elongation conformation to the initiation conformation to allow the cycle to resume. Termination at class II termination signals is enhanced by T7 lysozyme, which impedes the transition from an IC to an EC. For this reason, it had been suggested that some aspects of termination (at least at class II signals) may involve a reversal of the process that leads from an IC to an EC (9, 14, 34). The mechanism of termination at class I signals may differ, as termination at these signals is not affected by lysozyme. In support of this possibility, a number of mutants of T7 RNAP that affect termination at class II signals but not at class I signals have been identified. These mutations largely affect the flap domain, which interacts with the RNA–DNA hybrid at the upstream edge and are important for RNA displacement and the stability of the EC (7). In this work, we have observed that polymerases that are crosslinked in the EC configuration by the K179C–M750C disulfide bond appear to pause but are deficient in termination at a class II signal (Fig. 3). As noted in Fig. 1, K179 and M750 are located in the flap domain and specificity loop, respectively, whose interactions are critical to the formation of the RNA exit pore.
The different response of the RNAP to the class II pause site (the CJ signal) vs. the class II termination site (the PTH signal) is likely to depend on the stability of the complexes when halted in a U-rich context. Whereas complexes paused at the CJ signal (which lacks a U-rich region downstream from the conserved sequence) slowly resume transcription, most complexes halted at the PTH signal either terminate rapidly or proceed to further transcript extension. The observation that RNAPs that are crosslinked in the EC configuration exhibit similar kinetics of pausing and resumption of transcription at both the CJ and PTH signal indicates that the crosslink does not affect pausing, but stabilizes the complex during its transition through the U-rich region (resulting in a dramatically lowered efficiency of termination at the PTH signal). What might be the basis for this stabilization? As shown in Fig. 1, formation of the EC-specific crosslink involves elements in the specificity loop and the flap domain, which form part of the RNA exit pore and interact with the upstream edge of the RNA–DNA hybrid. Biochemical evidence suggests that conformational changes occur in the complex as it approaches the termination (release) site. These include collapse of the upstream edge of the transcription bubble and rearrangement of elements in the flap domain (24, 25). It seems likely that movement of these elements is involved in displacing or releasing the transcript during termination and that impeding the mobility or flexibility of these elements stabilizes the complex and makes termination a less likely outcome.
Recently, Mukherjee et al. (24) reported that a mutant T7 RNAP in which residues D660 and R379 had been substituted with Cys and allowed to form a crosslink exhibited a reduced efficiency of termination at a CJ-like class II signal. Because this effect was reversible upon reduction with DTT, it was concluded that alterations in the mobility of the two domains containing these residues (the finger and thumb domains, respectively) might be involved in termination. However, the position and orientation of these two residues in the IC and EC are nearly identical, and the significance of these findings relative to possible conformational changes associated with isomerization from an IC to an EC and reversal is unclear. It is possible that a transient movement of these elements occurs during the transition but that the relative positions of these elements before and after the transition remain the same.
The findings reported here may have relevance to the phenomenon of termination by multisubunit RNAPs. In general, two types of models for intrinsic termination by these RNAPs have been advanced. In one of these (the allosteric model) contact of the stem–loop structure in the RNA with the exit pore is thought to alter the structure of the enzyme, perhaps causing termination via an allosteric effect that is transmitted to the active site (35). In another type of model (rigid body), formation and growth of the stem–loop in the RNA competes with available space in the exit pore, and either pulls the RNA out of the complex (disrupting the upstream portion of the RNA–DNA hybrid) or pushes the complex forward, thereby disrupting the active site (36–38). In this work, we observed that, although formation of the K179C–M750C crosslink in T7 RNAP (which should restrict the mobility of elements in the exit pore) did not affect termination at a stem–loop class I terminator (Tφ), the crosslink reduced termination at a sequence-specific class II terminator (PTH). These results suggest that there are multiple pathways to termination by T7 RNAP, one of which requires flexibility of elements in the exit pore (sequence-specific or class II termination sites) and one of which does not (stem–loop or class I termination sites). If applied to observations with multisubunit RNAPs, the results obtained with T7 RNAP would favor a rigid body model for termination at intrinsic (stem–loop) termination signals.
Acknowledgments
We are grateful to Ray Castagna for technical assistance. This work was supported by National Institutes of Health Grant GM38147 (to W.T.M.).
Author contributions: K.M., D.T., M.A., and W.T.M. designed research; K.M. and D.T. performed research; K.M., D.T., M.A., and W.T.M. analyzed data; and K.M., D.T., M.A., and W.T.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: RNAP, RNA polymerase; IC, initiation complex; EC, elongation complex; T, template; NT, nontemplate; CJ, pause site for T7 RNAP in T7 concatemer junction; PTH, termination site for T7 RNAP found in human preparathyroid hormone gene.
References
- 1.McAllister, W. T. (1997) Nucleic Acids Mol. Biol. 11, 15–25. [Google Scholar]
- 2.Martin, C. T., Muller, D. K. & Coleman, J. E. (1988) Biochemistry 27, 3966–3974. [DOI] [PubMed] [Google Scholar]
- 3.Yin, Y. W. & Steitz, T. A. (2002) Science 298, 1387–1395. [DOI] [PubMed] [Google Scholar]
- 4.Tahirov, T. H., Temiakov, D., Anikin, M., Patlan, V., McAllister, W. T., Vassylyev, D. G. & Yokoyama, S. (2002) Nature 420, 43–50. [DOI] [PubMed] [Google Scholar]
- 5.Borukhov, S. & Nudler, E. (2003) Curr. Opin. Microbiol. 6, 93–100. [DOI] [PubMed] [Google Scholar]
- 6.Murakami, K. S. & Darst, S. A. (2003) Curr. Opin. Struct. Biol. 13, 31–39. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, M., Ma, N., Vassylyev, D. G. & McAllister, W. T. (2004) Mol. Cell 15, 777–778. [DOI] [PubMed] [Google Scholar]
- 8.Temiakov, D., Mentesana, P. E., Ma, K., Mustaev, A., Borukhov, S. & McAllister, W. T. (2000) Proc. Natl. Acad. Sci. USA 97, 14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macdonald, L. E., Durbin, R. K., Dunn, J. J. & McAllister, W. T. (1994) J. Mol. Biol. 238, 145–158. [DOI] [PubMed] [Google Scholar]
- 10.Jeng, S. T., Gardner, J. F. & Gumport, R. I. (1990) J. Biol. Chem. 265, 3823–3830. [PubMed] [Google Scholar]
- 11.Hartvig, L. & Christiansen, J. (1996) EMBO J. 15, 4767–4774. [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon, Y. S. & Kang, C. (1999) J. Biol. Chem. 274, 29149–29155. [DOI] [PubMed] [Google Scholar]
- 13.He, B., Kukarin, A., Temiakov, D., Chin-Bow, S. T., Lyakhov, D. L., Rong, M., Durbin, R. K. & McAllister, W. T. (1998) J. Biol. Chem. 273, 18802–18811. [DOI] [PubMed] [Google Scholar]
- 14.Lyakhov, D. L., He, B., Zhang, X., Studier, F. W., Dunn, J. J. & McAllister, W. T. (1998) J. Mol. Biol. 280, 201–213. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, X. & Studier, F. W. (2004) J. Mol. Biol. 340, 707–730. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, X. & Studier, F. W. (1997) J. Mol. Biol. 269, 10–27. [DOI] [PubMed] [Google Scholar]
- 17.Lyakhov, D. L., He, B., Zhang, X., Studier, F. W., Dunn, J. J. & McAllister, W. T. (1997) J. Mol. Biol. 269, 28–40. [DOI] [PubMed] [Google Scholar]
- 18.Brieba, L. G., Gopal, V. & Sousa, R. (2001) J. Biol. Chem. 276, 10306–10313. [DOI] [PubMed] [Google Scholar]
- 19.Dombkowski, A. A. & Crippen, G. M. (2000) Protein Eng. 13, 679–689. [DOI] [PubMed] [Google Scholar]
- 20.Temiakov, D., Tahirov, T. H., Anikin, M., McAllister, W. T., Vassylyev, D. G. & Yokoyama, S. (2003) Acta Crystallogr. D 59, 185–187. [DOI] [PubMed] [Google Scholar]
- 21.Temiakov, D., Anikin, M. & McAllister, W. T. (2002) J. Biol. Chem. 277, 47035–47043. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, M., Rong, M., Martin, C. T. & McAllister, W. T. (2001) J. Mol. Biol. 310, 509–522. [DOI] [PubMed] [Google Scholar]
- 23.Ma, K., Temiakov, D., Jiang, M., Anikin, M. & McAllister, W. T. (2002) J. Biol. Chem. 277, 43206–43215. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee, S., Brieba, L. G. & Sousa, R. (2003) EMBO J. 22, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn, Y. & Kang, C. (2005) Proc. Natl. Acad. Sci. USA 102, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mead, D. A., Skorupa, E. S. & Kemper, B. (1986) Protein Eng. 1, 67–74. [DOI] [PubMed] [Google Scholar]
- 27.Martin, F. H. & Tinoco, I. (1980) Nucleic Acids Res. 8, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheetham, G. M., Jeruzalmi, D. & Steitz, T. A. (1999) Nature 399, 80–83. [DOI] [PubMed] [Google Scholar]
- 29.Cheetham, G. M. & Steitz, T. A. (1999) Science 286, 2305–2309. [DOI] [PubMed] [Google Scholar]
- 30.Guo, Q., Nayak, D., Brieba, L. G. & Sousa, R. (2005) J. Mol. Biol. 353, 256–270. [DOI] [PubMed] [Google Scholar]
- 31.Theis, K., Gong, P. & Martin, C. T. (2004) Biochemistry 43, 12709–12715. [DOI] [PubMed] [Google Scholar]
- 32.Liu, C. & Martin, C. T. (2002) J. Biol. Chem. 277, 2725–2731. [DOI] [PubMed] [Google Scholar]
- 33.He, B., Rong, M., Durbin, R. K. & McAllister, W. T. (1997) J. Mol. Biol. 265, 275–288. [DOI] [PubMed] [Google Scholar]
- 34.Sousa, R., Patra, D. & Lafer, E. M. (1992) J. Mol. Biol. 224, 319–334. [DOI] [PubMed] [Google Scholar]
- 35.Toulokonov, I., Artsimovitch, I. & Landick, R. (2001) Science 292, 730–733. [DOI] [PubMed] [Google Scholar]
- 36.Gusarov, I. & Nudler, E. (1999) Mol. Cell 3, 495–504. [DOI] [PubMed] [Google Scholar]
- 37.Komissarova, N., Becker, J., Solter, S., Kireeva, M. L. & Kashlev, M. (2002) Mol. Cell 10, 1151–1162. [DOI] [PubMed] [Google Scholar]
- 38.Santangelo, T. J. & Roberts, J. W. (2004) Mol. Cell 14, 117–126. [DOI] [PubMed] [Google Scholar]
- 39.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]