Abstract
The spoligotyping method has become an important tool for the tracking of Mycobacterium tuberculosis strains in different epidemiological settings. In this study, we demonstrate the ability of the spoligotyping technique to accurately determine the pathogenetic mechanism of recurrent disease. This methodology has advantages over conventional restriction fragment length polymorphism methods which may be useful in large- scale intervention studies.
The direct repeat (DR) region in Mycobacterium tuberculosis is characterized by DR sequences interspersed with variable repeat sequences (8), which in combination have been termed DVRs (7). The DR region has been shown to be polymorphic in different clinical isolates of M. tuberculosis (8, 21). Polymorphism is generated by IS6110 insertion, deletion of DVR units by homologous recombination, and duplication of DVR repeat units by strand slippage during DNA replication (5, 19). The nature of polymorphism has been used to genotypically classify clinical isolates by DR restriction fragment length polymorphism (DR-RFLP) to define epidemiological relationships (14, 17). More recently, a PCR-based method was developed which has been termed spoligotyping (7, 9). This method can simultaneously identify M. tuberculosis and M. bovis species as well as provide a genotypic classification (9). Spoligotyping detects the presence (or absence) of 43 unique DVR repeat sequences by line-blot hybridization. Strains are differentiated on the basis of the presence of specific variable repeat sequences. Although it is widely accepted that the discriminatory power of the spoligotyping method is lower than that of the internationally standardized RFLP method (10), spoligotyping remains an important tool to genotype clinical isolates in different epidemiological settings (1, 3). These data have been used to monitor the spread of specific strains within defined geographical regions (15) and between different countries (16). The versatility of the spoligotyping method has also allowed certain nonepidemiological questions to be answered. The method was shown to be a useful tool for rapid identification of laboratory error and contamination (4, 12). When used in this context, the objective is to establish whether serial isolates collected from a single patient are genotypically identical or different. A finding of different genotypes is suggestive of contamination or laboratory error. Similarly, spoligotyping has been used to rapidly screen for genotypically identical isolates cultured on the same day, which is suggestive of laboratory contamination.
The interpretation of all molecular epidemiological data is based on establishing whether a strain is identical to or different from other strains found within a study community. Strains with shared genotypes are thought to represent ongoing transmission, while strains with unique genotypes are thought to represent reactivation (13). When this principle is applied to serial isolates collected from a single patient, it is possible to relate the genotype of the infecting strain to the genotype of a strain from a prior episode of disease (20). In this context, relapse is defined as the endogenous reactivation of the initial strain and, therefore, the strain genotype for each episode is identical. In contrast, reinfection is defined as a subsequent episode of disease resulting from exogenous infection with a different strain. Therefore, strains from the different episodes of disease will be genotypically different, provided that there is sufficient genotypic diversity within the M. tuberculosis population in the study setting. By investigating this important epidemiological question, it should be possible to determine the efficacy of treatment within different settings and also to identify factors influencing disease dynamics.
In this study, we have investigated whether the spoligotyping method is suitable for accurate identification of pathogenic mechanisms causing recurrent tuberculosis disease. Specimens from 38 patients with recurrent disease after curative treatment were investigated; these patients were drawn from a larger study of recurrent tuberculosis among gold miners in Free State, South Africa. All the patients completed treatment for the first disease episode under conditions of direct observed therapy. Eighteen patients were shown to be clinically cured, with tests showing both smear- and culture-negative results after 6 to 8 months of therapy, while isolates from the remaining 20 patients were smear negative at completion of therapy. On subsequent presentation, tuberculosis disease was diagnosed using the standard case definitions (2). Isolates from each episode were cultured on Lowenstein-Jensen medium, and the cells were resuspended in water and stored at −70°C for periods of up to 3 years. DNA was isolated from subcultures grown on Lowenstein-Jensen medium and genotypically characterized by IS6110 RFLP analysis using the internationally standardized protocol (18). The IS6110 banding patterns from isolates representing each episode of disease were compared, and isolates having different banding patterns were defined as representing cases of reinfection, while isolates having identical banding patterns were defined as representing cases of relapse (Fig. 1). In 16 patients, recurrence was classified as relapse, while in 21 patients, recurrence was classified as reinfection (Table 1). For patient 31, an RFLP result could not be obtained due to the presence of nontuberculous mycobacteria.
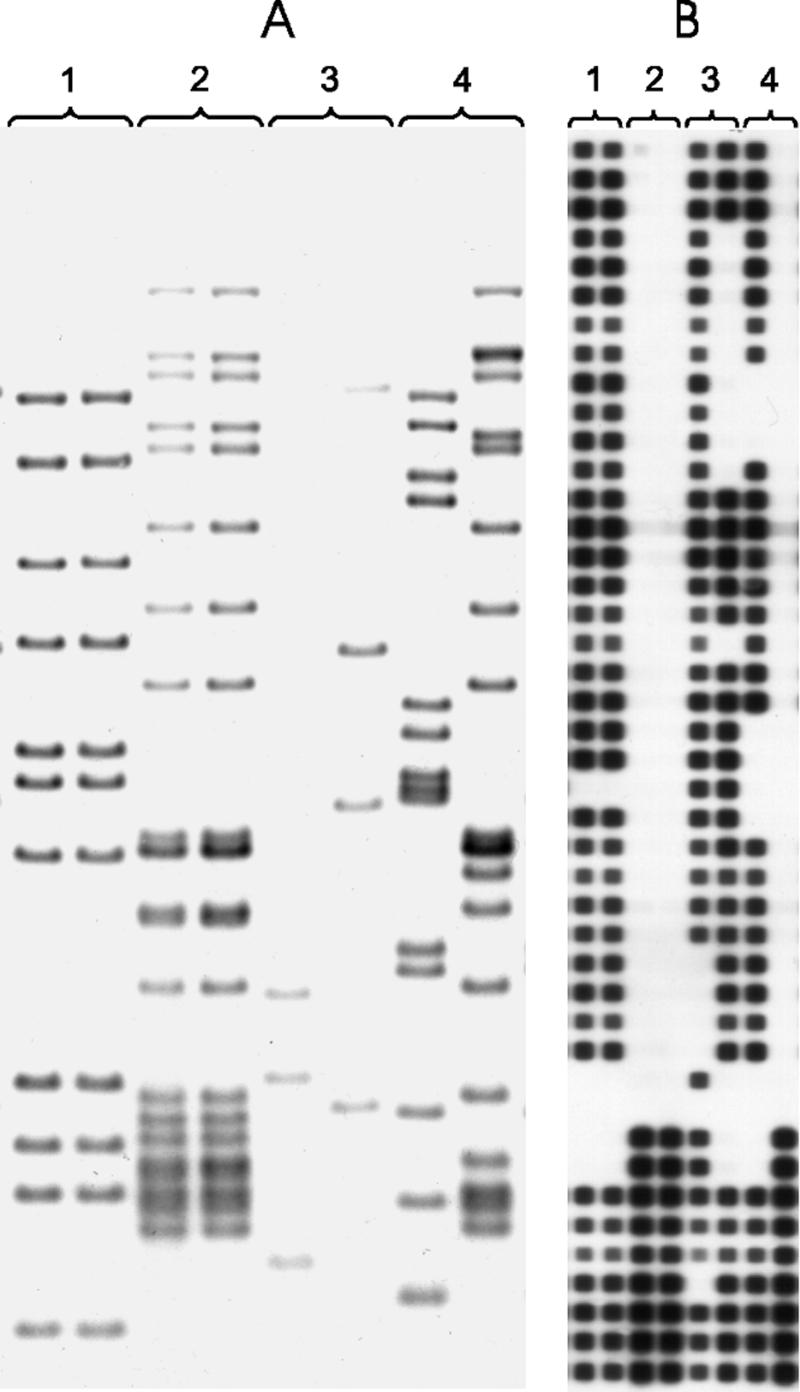
FIG. 1.
IS6110 RFLP and spoligotype analysis of clinical isolates collected from different episodes of disease from patients with recurrent disease. (A) IS6110 RFLP analysis. Pairs of lanes under a single lane number represent the isolates taken from a single patient but different episodes of disease. Lanes 1 and 2 show that the results for the two episodes represented in each pair of lanes are genotypically identical for each of those patients, demonstrating relapse, while lanes 3 and 4 show that the results for the two episodes represented in each pair of lanes are genotypically different for each of those patients, demonstrating reinfection. (B) Spoligotype analysis. Each pair of isolates represented corresponds to the isolates represented under the same lane number in panel A. Lanes 1 and 2 show that the results for the two episodes represented in each pair of lanes are genotypically identical for each of those patients, demonstrating relapse, while lanes 3 and 4 show that the results for the two episodes represented in each pair of lanes are genotypically different for each of those patients, demonstrating reinfection.
TABLE 1.
Genotypic classification of the pathogenic mechanism of recurrent M. tuberculosis disease by using IS6110 RFLP analysis and spoligotyping
| Patient | IS6110 RFLP
|
Spoligotyping
|
||
|---|---|---|---|---|
| Pattern | Mechanism | Pattern | Mechanism | |
| 1 | Different | Reinfection | Different | Reinfection |
| 2 | Different | Reinfection | Different | Reinfection |
| 3 | Same | Relapse/dual | Different | Reinfection |
| 4 | Different | Reinfection | Different | Reinfection |
| 5 | Different | Reinfection | Different | Reinfection |
| 6 | Different | Reinfection | Different | Reinfection |
| 7 | Same | Relapse | Same | Relapse |
| 8 | Same | Relapse | Same | Relapse |
| 9 | Different | Reinfection | Different | Reinfection |
| 10 | Same | Relapse | Same | Relapse |
| 11 | Different | Reinfection | Different | Reinfection |
| 12 | Different | Reinfection | Different | Reinfection |
| 13 | Different | Reinfection | Different | Reinfection |
| 14 | Different | Reinfection | Different | Reinfection |
| 15 | Same | Relapse | Same | Relapse |
| 16 | Evolveda | Relapse | Same | Relapse |
| 17 | Same | Relapse | Same | Relapse |
| 18 | Same | Relapse | Same | Relapse |
| 19 | Different | Reinfection | Different | Reinfection |
| 20 | Different | Reinfection | Different | Reinfection |
| 21 | Different | Reinfection | Different | Reinfection |
| 22 | Same | Relapse | Same | Relapse |
| 23 | Same | Relapse | Same | Relapse |
| 24 | Different | Reinfection | Different | Reinfection |
| 25 | Same | Relapse | Same | Relapse |
| 26 | Different | Reinfection | Different | Reinfection |
| 27 | Same | Relapse | Same | Relapse |
| 28 | Different | Reinfection | Different | Reinfection |
| 29 | Different | Reinfection | Different | Reinfection |
| 30 | Different | Reinfection | Different | Reinfection |
| 31 | No RFLP | Unknown | Different | Reinfection |
| 32 | Same | Relapse | Same | Relapse |
| 33 | Different | Reinfection | Different | Reinfection |
| 34 | Same | Relapse | Same | Relapse |
| 35 | Same | Relapse | Same | Relapse |
| 36 | Different | Reinfection | Different | Reinfection |
| 37 | Different | Reinfection | Different | Reinfection |
| 38 | Evolveda | Relapse | Same | Relapse |
Paired isolates show only minor changes in the IS6110 banding, suggesting evolution by transposition.
The DNA samples were then subjected to spoligotyping in a different laboratory. The recipient laboratory was blinded to the RFLP results. Spoligotyping was done using the internationally standardized method (9). All PCRs were prepared in specially designated areas (in laminar flow hoods) to prevent contamination by amplicons. Each series of PCRs included water blanks to identify possible reagent contamination. In addition, H37Rv DNA was included as a reference. No amplicon contamination was detected in any of the PCRs, and the H37Rv spoligotype pattern was consistent for each blot and matched the previously reported pattern (19), demonstrating repeatability of the technique. Figure 1 shows an example of the spoligotype pattern for isolates collected from patients with recurrent disease and the classification of relapse and reinfection.
From Table 1 it can be seen that in 36 of 37 (97%) cases, the spoligotype-defined mechanism of recurrence corresponded exactly to that obtained from IS6110 RFLP data. The results differed in only one case (patient 3), where IS6110 RFLP analysis had demonstrated an underlying dual infection in the initial infection. This resulted in the detection of a different spoligotype pattern in the second episode of disease. In the remaining patient (patient 31), IS6110 RFLP data were unable to provide a definition of the mechanism of recurrence, although the spoligotyping method classified the mechanism of recurrence as reinfection. A similar result was obtained when the DNA from isolates from two of the patients was spoligotyped (data not shown) and an RFLP classification could be obtained only after reculturing and DNA extraction. This demonstrates that the PCR-based spoligotype method is sufficiently robust to allow the amplification of the DR region even when there is insufficient DNA for RFLP analysis or the DNA is of a poor quality.
From the above results, the sensitivity and specificity of the spoligotyping method were calculated to be 100% and 94% (95% confidence interval, 82 to 100%), respectively. This suggests that the spoligotyping results correlate well to the standardized RFLP results when used to determine the pathogenetic mechanism of recurrent disease. This correlation demonstrates that there is sufficient genetic diversity within the DR locus of unrelated isolates (independent of IS6110 copy number) to allow the differentiation of strains. Furthermore, it is unlikely that such diversity is generated during persistent infection, as previous studies have shown that the stability of the spoligotype is higher than that of the IS6110 banding pattern (11). However, it is also acknowledged that this method is unable to distinguish dual infection in the first episode from relapse as a result of reactivation of only one of the initial infecting strains. As such dual infections are rare, it is unlikely that this will lead to a significant overestimate of the extent of reinfection. The spoligotyping method has numerous advantages over the standardized RFLP genotyping method in that it is better suited to high-throughput screening of patients in large intervention studies where relapse of tuberculosis must be distinguished from reinfection—for example, in evaluating different antituberculosis drug regimens. It has also been shown that this method can be adapted to genotypically classify strains directly from sputum (6). This would eliminate the need for the subculturing of each isolate for DNA isolation, thereby reducing the overall cost and exposure of laboratory workers to the pathogen. Furthermore, this method should greatly reduce the chance of laboratory contamination, which can result in the incorrect classification of the mechanism of recurrence.
Acknowledgments
We thank Tygerberg Hospital, the Harry Crossley Foundation, IAEA (projects SAF6/003 and CRP 9925), and Anglogold for financial assistance
We thank V. Moloi and D. Sehloho for the preparation and subculturing of tuberculosis specimens and M. Lekitlane for the maintenance of the tuberculosis database.
REFERENCES
- 1.Bauer, J., A. B. Andersen, K. Kremer, and H. Miorner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbett, E. L., G. J. Churchyard, T. Clayton, P. Herselman, B. Williams, R. Hayes, D. Mulder, and K. M. De Cock. 1999. Risk factors for pulmonary mycobacterial disease in South African gold miners. A case-control study. Am. J. Respir. Crit. Care Med. 159:94-99. [DOI] [PubMed] [Google Scholar]
- 3.Cronin, W. A., J. E. Golub, L. S. Magder, N. G. Baruch, M. J. Lathan, L. N. Mukasa, N. Hooper, J. H. Razeq, D. Mulcahy, W. H. Benjamin, and W. R. Bishai. 2001. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39: 3709-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de C. Ramos, M., H. Soini, G. C. Roscanni, M. Jaques, M. C. Villares, and J. M. Musser. 1999. Extensive cross-contamination of specimens with Mycobacterium tuberculosis in a reference laboratory. J. Clin. Microbiol. 37:916-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal, M., S. Lawn, B. Afful, J. W. Acheampong, G. Griffin, and R. Shaw. 1999. Spoligotyping in molecular epidemiology of tuberculosis in Ghana. J. Infect. 38:171-175. [DOI] [PubMed] [Google Scholar]
- 7.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 8.Hermans, P. W., D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemann, S., E. Richter, and S. Rusch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nivin, B., J. Driscoll, T. Glaser, P. Bifani, and S. Munsiff. 2000. Use of spoligotype analysis to detect laboratory cross-contamination. Infect. Control Hosp. Epidemiol. 21:525-527. [DOI] [PubMed] [Google Scholar]
- 13.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 14.Sola, C., L. Horgen, K. S. Goh, and N. Rastogi. 1997. Molecular fingerprinting of Mycobacterium tuberculosis on a Caribbean island with IS6110 and DRr probes. J. Clin. Microbiol. 35:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sola, C., A. Devallois, L. Horgen, J. Maisetti, I. Filliol, E. Legrand, and N. Rastogi. 1999. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg. Infect. Dis. 5:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sola, C., I. Filliol, M. C. Gutierrez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torrea, G., G. Levee, P. Grimont, C. Martin, S. Chanteau, and B. Gicquel. 1995. Chromosomal DNA fingerprinting analysis using the insertion sequence IS6110 and the repetitive element DR as strain-specific markers for epidemiological study of tuberculosis in French Polynesia. J. Clin. Microbiol. 33:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 21.van Soolingen, D., P. E. de Haas, J. Haagsma, T. Eger, P. W. Hermans, V. Ritacco, A. Alito, and J. D. van Embden. 1994. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J. Clin. Microbiol. 32:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]