Abstract
Poxviruses have evolved elaborate mechanisms for cell entry, assembly, and exocytosis. Recently, four vaccinia virus membrane proteins, namely A21, A28, H2 and L5, were reported to be necessary for cell entry and virus-induced cell–cell fusion but not for virion morphogenesis or attachment of virus particles to cells. Using immunoaffinity purification followed by mass spectrometry, we now show that these four proteins as well as four additional previously uncharacterized putative membrane proteins (A16, G3, G9, and J5) form a stable complex. These proteins fall into two groups: A21, A28, G3, H2, and L5 have an N-terminal transmembrane domain, 0–2 intramolecular disulfide bonds, and no sequence similarity, whereas A16, G9, and J5 have a C-terminal transmembrane domain and 4–10 predicted disulfide bonds and are homologous. Studies with conditional-lethal null mutants indicated that the viral membrane was crucial for assembly of the complex and that the absence of individual polypeptide components profoundly decreased complex formation or stability, suggesting a complicated interaction network. Analysis of purified virions, however, demonstrated that the polypeptides of the complex trafficked independently to the viral membrane even under conditions in which the complex itself could not be isolated. All eight proteins comprising the entry–fusion complex are conserved in all poxviruses, suggesting that they have nonredundant functions and that the basic entry mechanism evolved before the division between vertebrate and invertebrate poxvirus species.
Keywords: mass spectrometry, transmembrane proteins, vaccinia virus, viral envelope proteins, virus entry
Considerable progress has been made in determining the mechanisms of cell entry used by enveloped RNA viruses, but much less is known about this process for enveloped DNA viruses. Typically, RNA viruses have a single transmembrane glycoprotein that undergoes a conformational transition triggered by receptor recognition or low pH, leading to the insertion of a fusion peptide into the plasma membrane or the membrane of an endocytic vesicle (1, 2). Some RNA viruses, including members of the paramyxovirus family, have a variation of this theme in which separate proteins mediate attachment and fusion (3). The basic fusion machinery of the Herpesviridae, a family of large enveloped DNA viruses, includes three glycoproteins as well as subfamily-specific receptor binding proteins (4). Commensurate with their complexity, poxviruses have evolved more intricate entry mechanisms than other viruses (5). Two major infectious forms of vaccinia virus (VACV), the best-studied member of the Poxviridae, exist (6, 7). The mature virion (MV) assembles in cytoplasmic viral factories and can be released by cell lysis. However, some MVs are enveloped by modified trans-Golgi or endosomal cisternae to form doubly wrapped virions, which are transported along microtubules to the cell periphery, where loss of the outer of the two extra membranes occurs during exocytosis (7, 8). The resulting extracellular virion consists of a MV surrounded by one additional fragile membrane. MVs and extracellular virions attach to cells differently (9), in accord with the presence of different proteins in their respective surface membranes. However, a cellular receptor for either virion form has yet to be identified.
By using a reverse genetic approach, four MV transmembrane proteins (A21, A28, H2, and L5) were found to be required for entry of VACV (10–12). In each case, conditional-lethal null mutants produce MVs that appear morphologically normal but are noninfectious. These noninfectious MVs attach to cells, but their cores cannot penetrate into the cytoplasm. The same group of proteins is also required for virus-induced syncytia formation, indicating that they comprise part of the viral fusion machinery. Furthermore, the same MV proteins are required for cell-to-cell spread of extracellular virions, implying removal of the extra membrane before fusion. Evidence of physical interaction between A28 and H2 (10) raised the possibility that poxvirus entry–fusion proteins are organized into a multiprotein complex. Here we present an initial characterization of such a complex, show that it contains eight conserved proteins of which four were previously found to have a role in entry and membrane fusion, and demonstrate that formation of the viral membrane is necessary for assembly of the complex.
Materials and Methods
Recombinant Viruses. vLacOI encodes bacteriophage T7 RNA polymerase regulated by the Escherichia coli lac operator and E. coli lac repressor (13). vA11i (14), vA28i (15), vE10i (16), vA21i (11), vA16i (17), vG9i (S.O., unpublished work), and vFlagJ5i (S.O., unpublished work) are inducer-dependent derivatives of vLacOI containing the corresponding genes under the control of a bacteriophage T7 promoter and lac operator regulated by isopropyl β-d-thiogalactopyranoside (IPTG). vFlagJ5i encodes an inducible J5 protein with a Flag tag at its N terminus. Without IPTG addition, synthesis of the respective proteins and infectious virus formation are severely depressed. vA28-HAi/H2-V5 is a derivative of vA28i encoding A28 and H2 with C-terminal hemagglutinin epitope (HA) and V5 tags, respectively (10).
Antibodies to VACV Proteins. Anti-A28 antibody was obtained from rabbits immunized with a soluble portion of A28 protein expressed in insect cells (G.E.N., unpublished work); anti-A21, anti-L5, and anti-A16 polyclonal rabbit antibodies were produced against peptides from the corresponding proteins (11, 12, 17).
Infection and Transfection. BS-C-1 cells were infected with five plaque-forming units of VACV per cell or a recombinant virus in the presence or absence of 50 μM IPTG and harvested at ≈24 h. For transfection experiments, cells in six-well plates were infected and 1.5 h later were transfected with 1 μg of plasmid that had been incubated with 8 μg of Lipofectamine 2000 (Invitrogen).
Mass Spectrometry Analysis. Infected cells were disrupted by Dounce homogenization, nuclei were removed by centrifugation, and the supernatant was applied to a 36% sucrose cushion and centrifuged (18). The opalescent band of material floating at the top of the sucrose layer was found to be enriched in viral membrane proteins and was presumably derived from immature virions or their precursors (T.G.S., unpublished work). This membrane fraction was diluted 1:3 with PBS and concentrated by centrifugation for 1 h at 40,000 × g. The pellet was suspended by sonication in PBS/1% Nonidet P-40 (NP40); soluble material was incubated with agarose beads conjugated to anti-V5 antibody (Sigma) as described below. Bound proteins were eluted, resolved by SDS/PAGE and stained with Coomassie blue (GelCode Blue Stain Reagent, Pierce). Bands were excised from the polyacrylamide gel and digested with trypsin. Capillary liquid chromatography/tandem mass spectrometry and database searching were performed at the National Institute of Allergy and Infectious Diseases (NIAID) core facility.
Immunoaffinity Purification and Western Blot Analysis. Membrane proteins from whole infected cells, the membrane fraction described above, or sucrose gradient-purified virions were solubilized in PBS containing 1% NP40 for ≈20 min on ice with brief sonication. The extracts were clarified by centrifugation and incubated with unconjugated agarose beads for 1 h at 4°Cto remove material that nonspecifically bound agarose; they were further incubated at 4°C for 2 h or overnight with agarose beads conjugated to antibodies against corresponding epitope tags, V5 (Sigma), HA (Roche Applied Sciences), or Flag (Sigma). Agarose beads were washed extensively, and proteins were eluted in SDS/PAGE loading buffer, resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and analyzed with peroxidase-conjugated anti-HA rat monoclonal antibody (3F10, Roche Applied Sciences), anti-V5 mouse monoclonal antibody (Invitrogen), anti-Flag mouse monoclonal antibody (M2, Sigma), or polyclonal rabbit antibodies followed by anti-rabbit antibody conjugated to peroxidase and detected with a chemiluminescence detection kit (Pierce).
Analysis of free cysteines was carried out by using N-ethylmaleimide (NEM, Sigma) or 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS, Molecular Probes) as previously described (16).
Results
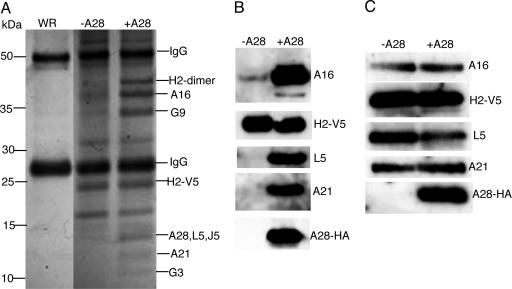
Identification of the Poxvirus Entry–Fusion Complex. We recently showed by coimmunoprecipitation that the H2 and A28 proteins interact in infected cells (10). Here we used immunoaffinity purification and mass spectrometry to find additional proteins associated with H2 and A28. Cells were infected with control wild-type VACV or recombinant virus vA28-HAi/H2-V5 (10), which inducibly expresses HA epitope-tagged A28 and constitutively expresses V5 epitope-tagged H2. A cytoplasmic fraction enriched in VACV membrane proteins was solubilized with NP40 nonionic detergent in PBS and incubated with immobilized antibody to the V5 tag. After extensive washing, the bound proteins were eluted, resolved by SDS/PAGE, and stained. As shown in Fig. 1A, numerous bands were resolved from the material derived from cells infected with vA28-HAi/H2-V5 in the presence of IPTG (i.e., allowing synthesis of A28-HA), whereas only bands corresponding to IgG heavy and light chains were seen clearly in the corresponding material from cells infected with the control virus that did not have an epitope tag. Furthermore, some of the specific bands stained less intensely or were not detected in the H2-V5 immunoaffinity-purified material from cells infected with vA28-HAi/H2-V5 in the absence of IPTG (i.e., not allowing synthesis of A28-HA) (Fig. 1 A).
Fig. 1.
Identification of viral proteins associated with a putative entry–fusion complex. (A) Cells were infected with vA28i-HA/H2-V5 in the presence (+A28) or absence (-A28) of IPTG or with control VACV (WR). Detergent-extracted proteins from a viral membrane-enriched fraction were bound to V5 antibody covalently linked to agarose beads and analyzed by SDS/PAGE. Protein bands stained with Coomassie blue dye are shown with the positions of mass markers on the left. Bands were excised from the sample with IPTG (+A28) and analyzed by mass spectrometry; the identified proteins are indicated on the right. (B) Immunoaffinity-purified samples with (+A28) and without (-A28) IPTG used in A were analyzed by Western blotting with antibodies to A16, A21, HA, L5, and V5 as indicated. (C) Virions from cells infected with vA28HAi/H2-V5 in the presence (+A28) or absence (-A28) of IPTG were purified by sucrose gradient sedimentation and analyzed by Western blotting with the antibodies used in B.
Protein bands in the sample from cells infected with vA28-HAi/H2-V5 in the presence of IPTG were excised from the gel and subjected to trypsin digestion followed by mass spectrometry analysis. In addition to monomer and dimer forms of H2, peptides from seven viral proteins, A16, A21, A28, G3, G9, J5, and L5, were identified in the indicated bands (Fig. 1 A). Each of these proteins has a predicted transmembrane domain, and three of them (A21, A28, and L5), in addition to H2, were previously shown to be required for cell entry and membrane fusion (10–12); a requirement of A16 for both processes was also determined recently (17). Although some of the proteins comigrated, precluding determination of their stoichiometry, they seemed to be present in roughly comparable amounts as judged by their intensity of staining relative to size. Learning the identities of the viral proteins in the stained bands helped us to interpret the results obtained in the absence of A28 synthesis. H2-V5 was present, because a V5 monoclonal antibody was used for immunoaffinity purification, but the A16, A21, A28, G3, G9, J5, and L5 proteins were absent or present in reduced amounts (Fig. 1 A), suggesting that A28 is important for complex formation or stability. A few other proteins not yet identified were present in the samples with and without IPTG, but their abundance relative to those specific for the sample with IPTG varied from experiment to experiment, and any significance to the isolation of those additional proteins with the complex remains to be determined.
The presence of a HA epitope tag on A28 and the availability of antibodies to A16, A21, and L5 allowed us to confirm the association of these four proteins with immunoaffinity-purified H2-V5 from the viral membrane fraction of cells infected with vA28-HAi/H2-V5 (Fig. 1B) as well as from detergent extracts of purified virions (data not shown). As anticipated from the stained gel in Fig. 1 A, the proteins A16, A21, A28, and L5 were not present or barely detected in the H2-V5 immunopurified material from cells infected in the absence of IPTG (Fig. 1B), despite the presence of these proteins in the membrane fraction of infected cells (data not shown) and purified virions (Fig. 1C). Thus, A28 was needed for formation of the complex but not for expression or viral membrane trafficking of at least four of the other component proteins.
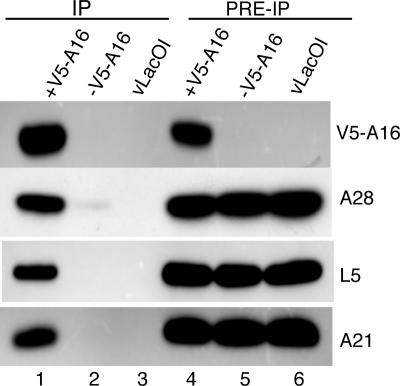
The copurification of at least seven proteins with H2 and the requirement for A28 could be explained by the existence of a single large complex. Alternatively, H2 and A28 might interact with the other proteins individually to form several small complexes. To investigate these possibilities, we carried out immunoprecipitations using epitope tags on other proteins of the putative complex. Cells were infected with vV5A16i, a recombinant VACV that inducibly expresses the A16 protein with an N-terminal V5 epitope tag (17), in the presence or absence of inducer or with vT7LacOI, the parent of vV5A16i. Analysis of solubilized cell extracts (Fig. 2, lanes 4–6) by Western blotting indicated that A21, A28, and L5 were expressed under all conditions but that synthesis of V5-tagged A16 was inducer-dependent in cells infected with vV5A16i. (A16 was not detected in extracts of cells infected with vT7LacOI because it does not have a V5 tag.) The proteins A21, A28, and L5 were captured along with V5-tagged A16 by antibody to V5 when V5A16 expression was induced but were barely detectable or undetectable in the absence of inducer (Fig. 2, lanes 1–3), confirming that they were in a complex with A16. In addition, the pattern of silver-stained bands seen on SDS/PAGE after affinity purification of the complex with V5 antibody from an extract of purified vV5A16i virions appeared similar to that in Fig. 1 A (data not shown). Furthermore, using a recombinant VACV expressing an inducible J5 with an N-terminal Flag tag, we found by Western blotting that A16, A21, A28, and L5 copurified with Flag-tagged J5 (data not shown). Consistent results were also obtained by using epitope tags associated with A21 and A28 (data not shown). Thus, no matter which protein of the complex was used for immunoaffinity purification, the other proteins in the group for which we had antibody were always detected.
Fig. 2.
Immunoaffinity purification of proteins associated with V5-tagged A16. Cells were infected with vV5A16i in the presence (+V5-tagged A16) or absence (-V5-tagged A16) of IPTG or with control, vLacOI. The NP40-soluble fraction of infected cells was analyzed by Western blotting before (PRE-IP) or after (IP) immunoaffinity purification with beads containing covalently bound antibody to the V5 epitope tag. The antibodies to A21, A28, L5, and V5 used for Western blotting are shown on the right.
Taken together, the mass spectrometry, SDS/PAGE, and Western blotting data indicated that each of the four proteins (A21, A28, H2, and L5) previously shown to be required for VACV entry and fusion as well as four additional previously uncharacterized proteins (A16, G3, G9, and J5) were present in a large complex.
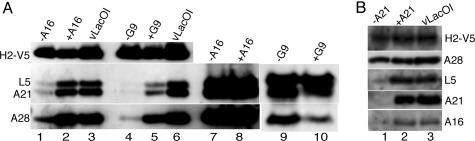
Contributions of Individual Subunits to Complex Formation. As shown in Fig. 1, the proteins of the putative entry–fusion complex were associated with H2 only in the presence of A28, suggesting that the latter protein is crucial for complex formation or stability. Additional inducible mutants were used to investigate complex formation in the absence of other individual proteins. We infected cells with vA16i in the presence or absence of IPTG or with the parental virus vLacOI and transfected the cells with DNA encoding H2-V5. (Note that the infected cells expressed endogenous nontagged H2, which presumably competed with the tagged version, thereby decreasing the amount of complex captured.) The proteins that coimmunoprecipitated with H2-V5 were analyzed by Western blotting. As shown in Fig. 3A, association of A21, A28, and L5 with H2-V5 occurred in cells infected with vLacOI or vA16i in the presence of IPTG, but the association of these proteins was greatly decreased when synthesis of A16 was repressed. Association of A21, A28, and L5 with H2-V5 was also greatly decreased when expression of G9 was repressed (Fig. 3A). In both cases, synthesis of each of the proteins was demonstrated by Western blotting of NP40-soluble cell extracts (Fig. 3A), indicating that protein complex formation was specifically perturbed by the absence of A16 or G9. Similar results were also obtained when H2 synthesis was repressed (data not shown).
Fig. 3.
Effects of the repression of genes encoding individual subunits on complex formation. (A) Cells were infected with vA16i in the absence (-A16) or presence (+A16) of IPTG or with vG9i in the absence (-G9) or presence (+G9) of IPTG or with vLacOI and transfected with a plasmid expressing H2-V5 controlled by its native promoter. NP40-soluble extracts were incubated with immobilized antibody to V5 (lanes 1–6), and the bound proteins were analyzed by Western blotting with monoclonal antibody to V5 (H2-V5) or with polyclonal antibodies to A21, A28, and L5 as indicated. As a control, 10% of the NP40-soluble extracts were analyzed directly by Western blotting (lanes 7–10). (B) Cells were infected with vA21i in the absence (-A21) or presence (+A21) of IPTG or with vLacOI and transfected with a plasmid expressing H2-V5 protein controlled by its native promoter. NP40-soluble extracts of infected cells were incubated with immobilized antibody to V5, and the bound proteins were analyzed as in A.
Results somewhat different from the above were obtained when synthesis of A21 was repressed. In the absence of A21, the amounts of L5 and A16 that coimmunoprecipitated with H2-V5 were greatly decreased, whereas the association of A28 with H2-V5 was hardly affected (Fig. 3B), suggesting a subcomplex. A21 associated with L5 was detected in the absence of A28, albeit in reduced amounts, but this complex was barely detected in the absence of H2 (data not shown). These observations suggest that the VACV entry–fusion complex has a complicated architecture, with individual subunits exerting multiple effects on the recruitment of other subunits and/or the stability of the complex.
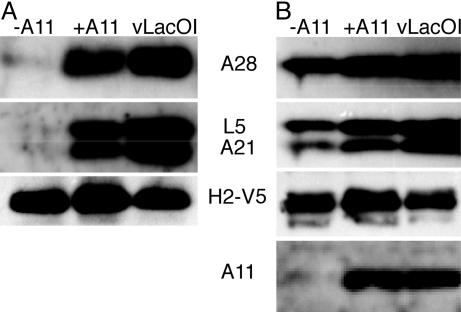
Viral Membranes Are Required for the Assembly of the Entry–Fusion Complex. Because VACV entry proteins are associated with the MV membrane, we were curious to know whether the complex would form in its absence, perhaps on cell membranes. To answer this question, we took advantage of conditional-lethal inducible mutant vA11i, which fails to induce viral membrane formation in the absence of IPTG because of repression of the nonvirion protein A11 (14). Cells infected with vA11i were transfected with DNA encoding H2-V5, and the proteins associated with H2-V5 were recovered by coimmunoprecipitation. The Western blot in Fig. 4A shows that A21, A28, and L5 were associated with H2-V5 when cells were infected with vT7LacOI or vA11i in the presence of IPTG. However, only trace amounts of these proteins were associated with H2-V5 when cells were infected with vA11i in the absence of IPTG (Fig. 4A). The proteins were detected, although in reduced amounts, in the NP40 soluble extracts used for immunoprecipitation (Fig. 4B). Thus, the viral membrane was required for the formation of the protein complex.
Fig. 4.
Requirement of viral membrane for complex formation. (A) Cells were infected with vA11i in the absence (-A11) or presence (+A11) of IPTG or with control vLacOI and transfected with a plasmid encoding H2-V5 protein controlled by its native promoter or mock-transfected (data not shown). NP40-soluble cell extracts were incubated with immobilized anti-V5 antibody, and bound proteins were resolved by SDS/PAGE and analyzed by Western blotting with antibodies to A28, L5, A21, and V5 as indicated. (B) As a control, ≈10% of NP40-soluble extracts were analyzed directly by Western blotting as in A.
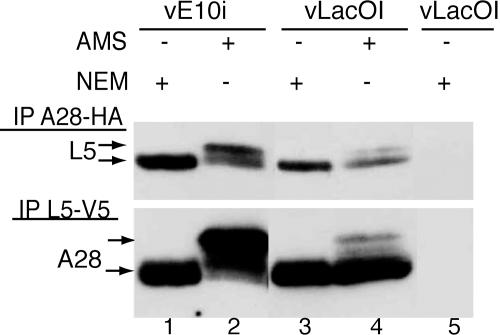
Intramolecular Disulfide Bonds Are Not Required for Formation of the Entry–Fusion Complex. With the exception of G3, the nonmembrane portions of the subunits of the entry–fusion complex contain multiple conserved cysteines. For A16 (17), A21 (11), A28 (19), H2 (T.G.S., unpublished work), and L5 (12), the cysteines were shown to form intramolecular disulfide bonds via the VACV-encoded redox pathway. It is known that repression of E10, the upstream component of the redox pathway, is required for virus maturation but not for viral membrane formation (16, 20). It was of interest, therefore, to determine whether E10 was needed for assembly of the entry–fusion complex. Cells were infected with vLacOI or vE10i, an inducible conditional lethal VACV (20), in the absence of IPTG and transfected with plasmids encoding A28-HA and L5-V5 proteins. Detergent-solubilized extracts were incubated with immobilized antibody to HA or V5, and the bound proteins were alkylated with N-ethylmaleimide (NEM) or AMS to determine the redox state of the cysteines. Each reduced cysteine that was alkylated by AMS increased the mass of the protein by ≈0.5 kDa, whereas alkylation by NEM had a negligible effect. First, we will consider the results obtained with cells infected with the control vLacOI. L5-V5 was captured with antibody to HA, and A28-HA was captured with antibody to V5, indicating that the proteins were associated in a complex. Furthermore, the mobilities of only trace amounts of the two proteins were changed by treatment with AMS (Fig. 5), indicating that the cysteines were disulfide-bonded. Association of L5-V5 and A28-HA also occurred in cells infected with vE10i in the absence of inducer, but the mobilities of the proteins increased with AMS, indicating that the cysteines were mostly in a reduced state (Fig. 5). Therefore, disulfide bond formation and virion maturation were not prerequisites for assembly of the entry–fusion complex.
Fig. 5.
Formation of entry–fusion complex with proteins lacking disulfide bonds. Cells were infected in the absence of IPTG with vE10i (lanes 1 and 2) or with vLacOI (lanes 3–5) and transfected (lanes 1–4) with plasmids encoding A28-HA and L5-V5 proteins controlled by their native promoters. NP40-soluble cell extracts were incubated with immobilized anti-HA (Upper) or anti-V5 (Lower) antibodies, and the bound proteins were treated with either N-ethylmaleimide (NEM) or AMS and analyzed by Western blotting with anti-V5 (Upper) or anti HA (Lower) antibodies.
Discussion
Progress in investigating poxvirus entry mechanisms depends on the identification of the proteins comprising the fusion complex. We have taken two approaches to identifying these proteins. The first approach involved reverse genetics, i.e., characterization of the phenotypes of conditional-lethal mutants. In this manner, we found that the A21, A28, H2, and L5 proteins were required for virus entry and cell–cell fusion (10–12). The second approach, used here, was based on the physical association of proteins. By epitope-tagging a protein implicated in entry and fusion by the reverse genetic approach, we were able to capture a complex containing additional proteins by immunoaffinity procedures. Seven viral proteins associated with H2 were identified by mass spectrometry. The presence of these proteins in a single complex was confirmed by Western blotting of proteins immunoaffinity-purified with epitope tags attached to several other proteins of this group, in addition to H2. This complex could be isolated after NP40 detergent treatment of whole infected cells, a membrane fraction enriched in viral proteins, or purified virions. Importantly, the four proteins identified by the reverse genetic approach were found in the complex. The four additional proteins were A16, G3, G9, and J5. A role in entry and fusion has now been demonstrated for A16 (17) and G9 (S.O., unpublished work). In addition, normal-looking virus particles were made in cells infected with a temperature-sensitive G3 mutant under nonpermissive conditions, consistent with a role for this protein in entry rather than virus assembly (S. E. M. Kato and R. Condit, personal communication). Although the function of the J5 protein has not been reported, the inability to isolate the corresponding deletion mutant suggests that it is essential (21). In addition to detecting J5 in the complex by mass spectrometry, we found that antibody to J5 could coimmunoprecipitate other proteins of the complex (unpublished work). Thus, functional data currently support roles in entry and fusion for at least six of the eight viral proteins in the complex.
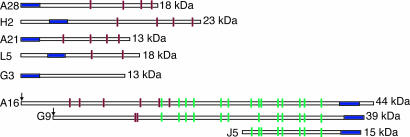
Structurally, the subunits of the poxvirus entry–fusion complex fall into two distinct groups: A21, A28, G3, H2, and L5 have an N-terminal transmembrane domain and 0–2 intramolecular disulfide bonds, whereas A16, G9, and J5 have a C-terminal transmembrane domain and have or are predicted to have 4–10 disulfide bonds (Fig. 6). The C-terminal segments of A21 (11), A28 (19), H2 (10), L5 (12), and, presumably, G3 are on the surface of the MV where they can interact with cell membrane components (Fig. 6). H2 and L5 have ≈30 aa N-terminal to the transmembrane domain, allowing for possible interactions with internal virion components, whereas the transmembrane domains of A21, A28, and G3 are located at the very N terminus (Fig. 6). A16, a member of the second group of entry–fusion proteins, has its N-terminal region on the MV surface (17), a transmembrane domain close to its C terminus, and a short C-terminal segment, which is presumably located within the virion (Fig. 6). The topologies of G9 and J5 are expected to be similar to that of A16, except that the transmembrane domains of G9 and J5 are located at the very C terminus. Both A16 and G9 are myristoylated (22). Despite their similar structural organization, the proteins of the first group have no detectable sequence similarity to each other (extensive comparisons notwithstanding) and therefore do not appear to be homologous. By contrast, the cysteine-rich domains of A16, G9, and J5 are homologous, suggesting that the respective genes evolved by duplications in the ancestral poxvirus genome (23).
Fig. 6.
Structural features of the components of the entry–fusion complex. Schematic drawings of the eight proteins are shown with transmembrane domains (blue) and masses indicated. Red ticks denote cysteine residues conserved in all orthologous proteins. Green ticks indicate cysteines that are conserved in orthologs and homologs. The N-terminal sites of the myristoylation of VACV proteins are indicated by arrows.
Preliminary experiments were carried out to investigate subunit interactions by isolating complexes from cells infected with null mutants. We hoped to obtain subcomplexes missing the binding partners of the respective proteins, but the absence of individual entry–fusion proteins greatly diminished the assembly or stability of the complex, suggesting a highly organized architecture. For example, no protein of the group was appreciably associated with H2 when A28 was absent, although the proteins were present in the viral membrane. However, at least one subcomplex containing A28 and H2 was detected in the absence of A21. Clearly, the subunit interactions need to be investigated by other methods. In experiments with vA11i, a conditional-lethal mutant that is defective in membrane biogenesis, no entry–fusion complex was detected, indicating that the viral membrane is required for the stable association of the complex subunits.
Of the eight proteins in the putative entry–fusion complex, five were shown to form intramolecular disulfide bonds via the VACV-encoded redox pathway (refs. 11, 12, 17, and 19 and T.G.S., unpublished work), and two more are predicted to form disulfides. Nevertheless, reduced forms of the entry–fusion proteins assembled to form a complex even when the redox pathway and late stages of morphogenesis were blocked by using a conditional-lethal mutant, vE10i. Nevertheless, the disulfide bonds are likely to be necessary for function and/or stability of the complex.
With the exception of G3, each protein component of the complex was known to have an ortholog in insect poxviruses as well as in all poxviruses of chordates with sequenced genomes (24). By screening entomopoxvirus protein sequences for similarity to chordopoxvirus G3 sequences with the psi-blast program, AMV071 from Amsacta moorei entomopoxvirus and MSV049 from Melanoplus sanguinipes entomopoxvirus were predicted to be orthologs (T.G.S., unpublished work). Conservation of the eight known components of the fusion protein complex throughout the poxvirus family strongly suggests that the basic entry mechanism arose early in their evolution and is largely unchanged. In view of the number of proteins involved in poxvirus entry, we anticipate that the fusion mechanism will differ appreciably from those of other, less complex, viruses.
Acknowledgments
We thank Eugene Koonin for help in sequence analysis and useful discussions and the National Institute of Allergy and Infectious Diseases (NIAID) mass spectrometry facility for protein analysis. This work was supported by the intramural program of the NIAID.
Author contributions: T.G.S. and B.M. designed research; T.G.S. and S.O. performed research; T.G.S., S.O., A.T., and G.E.N. contributed new reagents/analytic tools; T.G.S., S.O., and B.M. analyzed data; and B.M. and T.G.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: VACV, vaccinia virus; MV, mature virion; IPTG, isopropyl β-d-thiogalactopyranoside; HA, hemagglutinin epitope; NP40, Nonidet P-40; AMS, 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid.
References
- 1.Earp, L. J., Delos, S. E., Park, H. E. & White, J. M. (2005) Curr. Top. Microbiol. Immunol. 285, 25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sieczkarski, S. B. & Whittaker, G. R. (2005) Curr. Top. Microbiol. Immunol. 285, 1-23. [DOI] [PubMed] [Google Scholar]
- 3.Bagai, S. & Lamb, R. A. (1995) J. Virol. 69, 6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spear, P. G. & Longnecker, R. (2003) J. Virol. 77, 10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moss, B. (2006) Virology 344, 48-54. [DOI] [PubMed] [Google Scholar]
- 6.Moss, B. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott Williams & Wilkins, Philadelphia), 4th Ed., Vol. 2, pp. 2849-2883. [Google Scholar]
- 7.Smith, G. L., Vanderplasschen, A. & Law, M. (2002) J. Gen. Virol. 83, 2915-2931. [DOI] [PubMed] [Google Scholar]
- 8.Moss, B. & Ward, B. M. (2001) Nat. Cell Biol. 3, E245-E246. [DOI] [PubMed] [Google Scholar]
- 9.Vanderplasschen, A. & Smith, G. L. (1997) J. Virol. 71, 4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senkevich, T. G. & Moss, B. (2005) J. Virol. 79, 4744-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townsley, A., Senkevich, T. G. & Moss, B. (2005) J. Virol. 79, 9458-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Townsley, A., Senkevich, T. G. & Moss, B. (2005) J. Virol. 79, 10988-10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander, W. A., Moss, B. & Fuerst, T. R. (1992) J. Virol. 66, 2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Resch, W., Weisberg, A. S. & Moss, B. (2005) J. Virol. 79, 6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senkevich, T. G., Ward, B. M. & Moss, B. (2004) J. Virol. 78, 2357-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senkevich, T. G., White, C. L., Koonin, E. V. & Moss, B. (2000) Proc. Natl. Acad. Sci. USA 97, 12068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ojeda, S., Senkevich, T. G. & Moss, B. (2006) J. Virol. 80, 51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earl, P. L., Cooper, N., Wyatt, S., Moss, B. & Carroll, M. W. (1998) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 2, pp. 16.16.1-16.16.3. [Google Scholar]
- 19.Senkevich, T. G., Ward, B. M. & Moss, B. (2004) J. Virol. 78, 2348-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senkevich, T. G., Weisberg, A. & Moss, B. (2000) Virology 278, 244-252. [DOI] [PubMed] [Google Scholar]
- 21.Zajac, P., Spehner, D. & Drillien, R. (1995) Virus Res. 37, 163-173. [DOI] [PubMed] [Google Scholar]
- 22.Martin, K. H., Grosenbach, D. W., Franke, C. A. & Hruby, D. E. (1997) J. Virol. 71, 5218-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senkevich, T. G., Koonin, E. V., Bugert, J. J., Darai, G. & Moss, B. (1997) Virology 233, 19-42. [DOI] [PubMed] [Google Scholar]
- 24.Upton, C., Slack, S., Hunter, A. L., Ehlers, A. & Roper, R. L. (2003) J. Virol. 77, 7590-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]