Abstract
A new strain of Ibaraki virus (IBAV) was isolated from cattle showing atypical symptoms of Ibaraki disease. The isolate was genetically characterized, and the genetic diversity and evolution of the capsid proteins of viruses in the epizootic hemorrhagic disease virus (EHDV) serogroup were investigated. The nucleotide sequences of the isolate's viral RNA segments 2, 3, 6, and 7, which encode the viral structural proteins VP2, VP3, VP5, and VP7, respectively, were determined and were then compared against those of the existing strains of IBAV and EHDV, to which IBAV belongs serologically. The nucleotide sequences of segments 3 and 7 were conserved within the EHDV serogroup, particularly well among the strains of IBAV and Australian EHDV. The similarity of the sequence of segment 6 of the isolate to sequences of corresponding segments of the other strains of IBAV and EHDV was found to be about 93%. The similarity of segment 2 of the isolate to segments 2 of the other strains of IBAV and EHDV was less than 70%. Phylogenetic analysis based on the deduced amino acid sequences of segments 3 and 7 revealed that the viruses differed according to their geographical distributions. However, the new isolate of IBAV was categorized as having a distinct lineage in the phylogenetic tree of VP2. These results suggest that the isolate was modified by a reassortment of segment 2 and that it exhibits unique genetic and antigenic characteristics.
Ibaraki virus (IBAV) is a member of the epizootic hemorrhagic disease virus (EHDV) serogroup in the genus Orbivirus, family Reoviridae. Although IBAV is distinguishable from the eight other serotypes of EHDV within the serogroup, a partial cross-neutralization between IBAV and EHDV serotype 2 (EHDV-2) has been found (3, 6, 29). IBAV is transmitted by Culicoides sp. biting midges and causes Ibaraki disease, which is characterized by fever, anorexia, and a deglutitive disorder in cattle (22, 23). The disease was initially reported in Japan in 1959, and since then there have been epidemics of it in east Asia (1, 17) and antibodies to the virus have been found in bovine sera in Australia and Indonesia (5, 10, 18).
In the late summer to autumn of 1997, a large-scale epidemic of Ibaraki disease occurred in western Japan. During the same period of time, numerous miscarriages and stillbirths occurred in apparently healthy cattle in the same area. In the 1997 epidemic, a new strain of IBAV was isolated not only from cattle showing Ibaraki disease but also from aborted fetuses, their dams, and biting midges, as well as from the blood of both affected and apparently healthy cattle. The new virus was distinguished from the existing strains of IBAV by a cross-neutralization test and by PCR-restriction fragment length polymorphism analysis (21).
The IBAV genome is composed of 10 double-stranded (ds) RNA segments in a bilayered capsid. Segments 3 and 7 encode the inner capsid proteins VP3 and VP7, respectively. These capsid proteins play the role of serogroup-specific antigens. Segments 2 and 6 encode the outer layer proteins VP2 and VP5, respectively, of which VP2 is a major neutralizing antigen and serotype-specific determinant (14). The 10 genomic segments have the potential to be exchanged within the serogroup (reassortment). In another orbivirus, bluetongue virus (BTV), reassortment events in tissue culture, in the vector, and in nature have been described (7, 9, 26). The molecular characterization of IBAV would be valuable for investigating the viral evolution and epidemiology of the disease.
In this report, four RNA segments of IBAV, including the new strain, were sequenced and compared with the published sequence data in order to demonstrate the genetic heterogeneity within the EHDV serogroup, along with the consequences of virus evolution.
MATERIALS AND METHODS
Viruses and cells.
Propagation of the viruses in this study was done as previously described (21). The Ibaraki-2, Y87061, and KSB-14/E/97 IBAV strains, isolated in 1959, 1987, and 1997, respectively, and the CSIRO439 strain of Australian EHDV-2 were propagated on baby hamster kidney (BHK-21) cells. The cells were maintained in Eagle's minimum essential medium (Nissui Pharmaceutical Co., Tokyo, Japan) supplemented with 0.295% tryptose phosphate broth (Difco Laboratories, Detroit, Mich.), 0.15% sodium bicarbonate, 2 mM l-glutamine, and 5% calf serum.
Extraction of viral dsRNAs.
Viral dsRNAs were extracted from the infected BHK-21 cells as described previously (28), and the individual RNA segments were separated on a 0.8% agarose gel. The separated viral dsRNAs were excised from the agarose gel and purified with the RNaid kit (Bio 101, La Jolla, Calif.) in accordance with the manufacturer's instructions.
Genomic amplification and cloning.
The primers corresponding to the 5′ and 3′ termini of segments 2, 3, 6, and 7 were synthesized on the basis of the sequences of the No.2 strain of IBAV (Table 1) (15, 19, 20). Reverse transcription-PCR (RT-PCR) was performed with the Titan One Tube RT-PCR kit (Roche Diagnostics, Indianapolis, Ind.) containing a proofreading polymerase. The RT-PCR mixture consisted of 0.2 mM (each) deoxynucleoside triphosphate, 5 mM dithiothreitol, 1× RT-PCR buffer, 1.5 mM MgCl2, and an enzyme mixture in a total reaction volume of 50 μl. The purified viral RNA segment and both strand primers were incubated at 94°C for 5 min and quenched on ice prior to the addition of the reaction mixture. The RT was conducted at 48°C for 60 min. This was followed by an initial 3-min denaturation at 95°C and 10 subsequent cycles of denaturation at 95°C for 30 s, annealing at 55 to 63°C (depending on the primer pair) for 30 s, and elongation at 68°C for 2 min, and then by 25 cycles of denaturation at 95°C for 30 s, annealing at 55 to 63°C for 30 s, and elongation at 68°C for 2 min at first, though with each cycle the elongation time was incremented by 5 s.
TABLE 1.
Oligonucleotide primers used for the cDNA amplification and sequencing
| Primer | Sequence (5′→3′) | Position | Purpose |
|---|---|---|---|
| L2-1 | GTTAAATTGTTCCCAGAATGGAGG | 1-24 | cDNA amplification |
| L2-6 | GTAAGTTTGTTGTTCCCAGTAA | 2981-3002 | cDNA amplification |
| IBAL2R | GTAAGTTTGTTGTTCCCAGTAAACC | 2978-3002 | cDNA amplification |
| L3F | GTTAAATTTCCAGAGCGATGGCG | 1-23 | cDNA amplification |
| L3R | GTAAGTGTATTTCCAGTGCTTTTCC | 2744-2768 | cDNA amplification |
| IBAM5F | GTTAAAAAGGAGGCAC GTTCTTGC | 1-25 | cDNA amplification |
| IBAM5R | GTAAGTGTAAGGAGTTCGCGTACTC | 1617-1641 | cDNA amplification |
| IBAVS7F | GGTTAAAATTTGGTGAAAATGG | 1-22 | cDNA amplification |
| IBAVS7R | GTAAGTTGAATTTGGGAAAACG | 1141-1162 | cDNA amplification |
| M13Forward | CACGACGTTGTAAAACGAC | Sequencing | |
| M13Reverse | GGATAACAATTTCACACAGG | Sequencing | |
| IBAL2552F | GTTTGCATTGGCTCAGGAAC | 552-571 | Sequencing |
| IBAL22465R | CGCCATCCTGCATGATATTTC | 2227-2246 | Sequencing |
| IBAL21906F | CAAGTGTAACTATCATTACG | 1906-1925 | Sequencing |
| IBAL32233R | ACTCGATCGATAGCTGGAGG | 2214-2233 | Sequencing |
| IBAL3557F | GGTGCAGCATATATTAGACGG | 557-576 | Sequencing |
| IBAL31165F | CCAAAATATGGCACGACGACAGC | 1165-1184 | Sequencing |
| IBAM5IF | GTCTTATGGCAAAGTGATTGG | 495-515 | Sequencing |
| IBAM5IR | GCCAGCAGCCACAAATTCAT | 829-848 | Sequencing |
The purified PCR products were ligated into pGEM-T Easy Vector systems (Promega, Madison, Wis.). The recombinant plasmids were transfected into competent Escherichia coli JM 109 cells. The positive clones were confirmed by PCR using M13 forward and reverse primers. A plasmid containing the viral genome was obtained with a commercial plasmid extraction kit (Promega) according to the manufacturer's protocol.
Sequencing.
The sequencing primers are shown in Table 1. Three individual clones containing the viral genome were sequenced in both directions by using a model 4200 automated DNA sequencer (Li-Cor Inc., Lincoln, Nebr.) with the Thermo Sequenase cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio).
Genetic analysis.
The nucleotide sequences obtained in this study were edited with the GENETYX software. The multiple alignments of the deduced amino acid sequences encoded by the open reading frames (ORF) were done with the CLUSTAL W program (31). The aligned sequences were used to construct the phylogenetic trees by the neighbor-joining method (27), and the distances were corrected by using Kimura's two-parameter method (16). The reliability of the branching orders was estimated by bootstrapping (1,000 samples). The phylogenetic trees were drawn with TreeView software (24).
Sequence data.
The sequence data determined in this study and previously reported nucleotide sequences of IBAV and Australian and North American EHDV segments 2, 3, 6, and 7 were used in the genetic analysis. The sequence data were taken from the nucleotide database (the accession numbers are shown in the legends for Fig. 1 and 2).
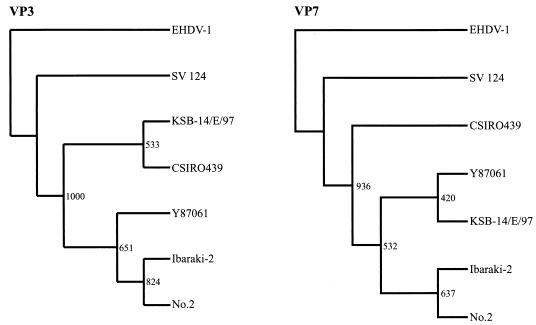
FIG. 1.
Phylogenetic tree based on VP3 and VP7 of IBAV and EHDV. Trees were constructed by the neighbor-joining method and bootstrap test (n = 1,000). EHDV-1 was used as an outgroup to root the tree. Accession numbers are as follows: EHDV-1 segment 3, X61589; EHDV-1 segment 7, D10766; SV 124 (EHDV-2) segment 3, L33819; SV 124 segment 7, AF188643; CSIRO439 (EHDV-2) segment 3, S68010; No.2 (IBAV) segment 3, AB04930; No.2 segment 7, AB041934.
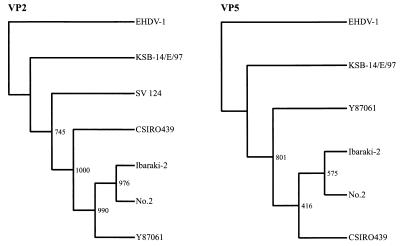
FIG. 2.
Phylogenetic tree based on VP2 and VP5 of IBAV and EHDV. Trees were constructed by the neighbor-joining method and bootstrap test (n = 1,000). EHDV-1 was used as an outgroup to root the tree. Accession numbers are as follows: EHDV-1 segment 2, D10767; EHDV-1 segment 6, X55782; SV 124 (EHDV-2) segment 2, L33818; No.2 (IBAV) segment 2, AB030735; No.2 segment 6, AB030736.
Nucleotide sequence accession numbers.
The IBAV and Australian EHDV-2 genome sequences reported in this paper have been deposited in the DDBJ, EMBL, and GenBank databases and given the following accession numbers: AB078620 through AB078633.
RESULTS
Sequence determination and analysis.
cDNA clones were generated for segments 2, 3, 6, and 7 of the strains Ibaraki-2, Y87061, and KSB-14/E/97 of IBAV and for segments 2 and 6 of the strain CSIRO439 of EHDV-2. To reveal the phylogenetic relationships within the EHDV serogroup, these clones were sequenced, and the sequence data were compared against the published data for strain No.2 of IBAV, strain CSIRO439 of Australian EHDV-2, strains SV 123 and Alberta of North American EHDV-1, and strain SV 124 of North American EHDV-2.
The ORFs of segments 3 and 7 of all the strains of IBAV comprised 2,700 and 1,050 nucleotides, respectively. The variations of segments 3 and 7 within the strains of IBAV and CSIRO439 ranged from 92.8 to 93.7% for segment 3 and from 92.6 to 94.1% for segment 7, whereas the variations between segments 3 and 7 of IBAV together with CSIRO439 and those of SV 124 of EHDV-2 were both approximately 80% (Table 2). The deduced amino acid sequences of VP3 and VP7 were relatively conserved at more than 99% for the IBAV strains, except for the VP7 sequences of strain No.2. The amino acid sequence identities for VP7 between IBAVs and CSIRO439 ranged from 96.0 to 98.9%. The VP3 and VP7 amino acid sequence identities between IBAVs and SV 124 of EHDV-2 ranged from 95.3 to 95.9% for VP3 and from 94.6 to 97.1% for VP7. Similar lower identities between the VP3 and VP7 proteins of CSIRO439 and those of SV 124 were observed.
TABLE 2.
Nucleotide and deduced amino acid sequence identities of segments 2, 6, 3, and 7 among IBAV and EHDV strains
| Segment and virus | % Identitya to indicated strain of:
|
|||||
|---|---|---|---|---|---|---|
| IBAV
|
EHDV-2
|
|||||
| No.2 | Ibaraki-2 | Y87061 | KSB-14/E/97 | CSIRO439 | SV 124 | |
| Segment 2 | ||||||
| No.2 | 98.5 | 96.5 | 69.4 | 94.5 | 70.9 | |
| Ibaraki-2 | 97.8 | 97.4 | 69.7 | 95.3 | 71.2 | |
| Y87061 | 97.4 | 98.5 | 69.9 | 94.4 | 71.4 | |
| KSB-14/E/97 | 67.5 | 67.9 | 68.2 | 69.8 | 68.7 | |
| CSIRO439 | 95.2 | 96.2 | 96.4 | 68.4 | 71.7 | |
| SV 124 | 72.4 | 73.3 | 73.8 | 67.4 | 73.8 | |
| Segment 6 | ||||||
| No.2 | 99.4 | 97.1 | 92.6 | 93.5 | ||
| Ibaraki-2 | 98.3 | 97.5 | 92.9 | 93.9 | ||
| Y87061 | 98.3 | 99.2 | 92.2 | 93.4 | ||
| KSB-14/E/97 | 97.3 | 98.3 | 98.7 | 93.1 | ||
| CSIRO439 | 97.7 | 98.7 | 98.7 | 98.1 | ||
| Segment 3 | ||||||
| No.2 | 97.8 | 97.7 | 93.6 | 92.8 | 80.5 | |
| Ibaraki-2 | 99.8 | 99.9 | 94.9 | 93.7 | 80.9 | |
| Y87061 | 99.7 | 99.9 | 94.8 | 93.6 | 80.8 | |
| KSB-14/E/97 | 99.0 | 99.2 | 99.3 | 92.8 | 79.8 | |
| CSIRO439 | 98.4 | 98.7 | 98.8 | 98.3 | 79.8 | |
| SV 124 | 95.6 | 95.8 | 95.9 | 95.3 | 94.9 | |
| Segment 7 | ||||||
| No.2 | 98.2 | 96.2 | 95.7 | 92.6 | 79.9 | |
| Ibaraki-2 | 97.1 | 97.8 | 97.3 | 94.1 | 81.0 | |
| Y87061 | 97.1 | 99.4 | 98.4 | 93.5 | 81.0 | |
| KSB-14/E/97 | 97.1 | 99.4 | 100 | 92.8 | 81.5 | |
| CSIRO439 | 96.0 | 98.3 | 98.9 | 98.9 | 80.7 | |
| SV 124 | 94.6 | 96.6 | 97.1 | 97.1 | 96.3 | |
Percentages of nucleotide identity are in lightface; percentages of deduced amino acid identity are in bold face.
The ORFs of segments 2 and 6 of Ibaraki-2, Y87061, and KSB-14/E/97 of IBAV and CSIRO439 of EHDV-2 were 2,949 and 1,015 nucleotides in length, respectively. The genomic sequence identities of segment 6 among the strains of IBAV and CSIRO439 of EHDV-2 were between 93.1 and 93.9%. The variations of segment 6 among IBAV strains No.2, Ibaraki-2, and Y87061 were under 2.9%. However, the identities of segment 6 between strain KSB-14/E97 and the other strains of IBAV were 92.2 to 92.6%, indicating a higher degree of genetic change than seen in segments 3 and 7. The deduced amino acid sequences of VP5s of the strains of IBAV and EHDV-2 had relatively high identities, ranging from 97.3 to 99.2%.
A remarkable genomic variation was observed on the nucleotide sequence of segment 2. In contrast with the identities (between 94.4 and 98.5%) of segments 2 of strains No.2, Ibaraki-2, Y87061, and CSIRO439, the identities of segments 2 of KSB-14/E/97 and these strains were less than 70%. Poor identities of the deduced amino acid sequence of KSB-14/E/97 VP2 with those of the other strains of IBAV and EHDV-2 were observed, and the identities had lower values than those between the VP2 sequences of SV 124 and the other strains of IBAV.
Phylogenetic analysis.
The phylogenetic trees of VP2, VP3, VP5, and VP7, constructed by using the deduced amino acid sequences encoded by the complete ORFs of segments 2, 3, 6 and 7, were individually compared. In the tree based on the VP3 sequence data (Fig. 1) there were two major clusters. SV 124 of EHDV-2 constituted an individual cluster, and the other cluster was composed of strains of IBAV and CSIRO439 of EHDV-2. Because strain SV 124 was isolated in North America and the other strains were isolated in Asia and Australia, the separation of these clusters seemed to correspond to the geographical distribution of the virus strains. The phylogenetic tree of VP7 resembled that of VP3 in its cluster construction. The clusters in the tree of VP7 were divided into three groups by geographical distribution and might define their geographical gene pool.
The phylogenetic trees of VP5 and VP2 were rather complicated. In the tree of VP5, strain KSB-14/E/97 was separated into a separate cluster (Fig. 2). However, strain CSIRO439 was in the same cluster as the existing strains of IBAV. The phylogenetic difference of strain KSB-14/E/97 was demonstrated more clearly in the tree of VP2. Strains No.2, Ibaraki-2, and Y87061 of IBAV were all in the same cluster, which was distinguishable both from the cluster of KSB-14/E/97 and from the cluster of SV 124 (Fig. 2).
DISCUSSION
IBAV is one of the most important insect-borne viruses in Japan. Since the first recognition of Ibaraki disease in Japan in 1959, epidemics have occurred in 1960, 1982, and 1987. However, reproductive problems in cows caused by IBAV were not reported until 1997. The serological tests and restriction fragment length polymorphism analysis of segment 3 of the 1997 isolate revealed that this new isolate belonged to IBAV but was distinguishable from the IBAVs isolated during past epidemics. This newly identified virus was considered a variant of IBAV (21). However, genetic characterization of this IBAV had not been performed, and its phylogenetic relationship with other EHDVs, as well as with other strains of IBAV, was unknown. In this study, we determined the sequences of the ORF regions of the four structural protein genes of this new variant virus and compared its nucleotide and deduced amino acid sequences with those of IBAV and EHDV in order to define the phylogenetic relationship of the virus.
The virus strains selected differed in geographical origin: the IBAV isolate was from Japan, and one EHDV-2 strain was from Australia, and the other was from North America. We compared sequences of segments 3 and 7, encoding serogroup-specific proteins VP3 and VP7, respectively, from the EHDV serogroup, to one another. The deduced amino acid sequences of the proteins were conserved, having 5% variability, between IBAV and EHDV. In the nucleotide sequences, there is a 20% variability between IBAV and North American EHDV, while there is a less than a 7% difference between IBAV and Australian EHDV. The phylogenetic analysis of VP3 showed two distinct clusters between the strains of IBAV and Australian EHDV-2. However, the KSB-14/E/97 strain of IBAV formed a single cluster with the CSIRO439 strain of EHDV-2. The result suggested that IBAV and Australian EHDV share a genetic pool. However, within the EHDV serogroup, phylogenetic analysis of segment 7 produced a more accurate geographical separation than analysis of segment 3, and so North American EHDV might be distinguishable from IBAV and Australian EHDV. Gould et al. demonstrated that a feature of the phylogenic trees based on VP3 within the EHDV and BTV serogroups could be clearly delineated on the basis of their geographical origins (11, 12, 25), and phylogenetic analyses revealed that the BTV segment 7 genes do not reflect the geographic origins of the virus strains (2, 32). Unlike the phylogenetic analyses of BTV, the phylogenetic analysis of IBAV and EHDV based on VP7 showed more clearly a geographical distribution. The VP7 has been identified as a protein that binds to vector cells (30, 33). The results of the phylogenetic analysis based on VP7 in this experiment, which divided the viruses into three groups (the North American and Australian EHDV-2 and the IBAV groups), might indicate viral adaptability to vector insects.
The nucleotide sequences of segments 6 from the strains of IBAV and CSIRO439 were relatively divergent, as were the sequences of segments 3 and 7. The identities of the deduced amino acid sequences of VP5 among these viruses were more than 98.1% and might bring the same antigenicity. Although the biological functions of VP5 are not well understood, VP5 was involved in the determination of virus serotype, possibly by imposing conformational constraints on VP2. The higher identities of VP5s from KSB-14/E/97 and the other strains of IBAV and Australian EHDV-2 might give a consistent explanation of the partial cross-reactivity of KSB-14/E/97 to other viruses in the cross-neutralization tests.
Segment 2 displayed remarkable genetic heterogeneity within the EHDV serogroup. The nucleotide sequence identities of segments 2 from KSB-14-E/97 and the existing strains of IBAV were less than 70%, and those of segments 2 from the existing strains of IBAV and the North American EHDV were lower than that. VP2 is the main neutralization antigen of the virus and is the determinant of virus serotype. The low identities of VP2 of KSB-14/E/97 might result in low reactivity in cross-neutralization tests with existing strains of IBAV. The phylogenetic analysis of VP2 and VP5 revealed that KSB-14/E/97 was segregated in a cluster distinct from that of IBAV and Australian EHDV-2. This phylogenetic distinction was remarkable in the tree of VP2. The greatest antigenic and sequence variations are also those exhibited by VP2s of BTV and EHDV (4, 8, 13). It is considered that such low nucleotide identity is the result of reassortment rather than the result of cumulative genetic mutation. The findings of the great variation of VP2, together with the findings of the variations of VP3, VP5, and VP7 of KSB-14/E/97, suggested that this new variant of IBAV was generated by a reassortment event in segment 2 and by mutations, including base substitution, deletion, and insertion, in segments 3, 6, and 7.
In natural conditions, reassortment occurs given the existence of at least two different strains present at the same time, in the same area, and in the same affected host. Although we could not estimate the other parental virus, the new variant of IBAV may have been generated in nature by genetic reassortment between IBAV and another orbivirus.
Our data analyzing four structural protein genes indicated that IBAV was clearly segregated from the North American EHDV and showed a slight difference from the Australian EHDV in nucleotides and amino acid sequences. It may be possible to regard IBAV and Australian EHDV-2 as nearly identical topotypes. Little is known about the mechanisms that induce such a variation between IBAV and Australian EHDV. When orbivirus infects host cells, VP2 is responsible for the attachment to mammalian cells, while VP7 mediates the attachment to insect cells. Therefore, vertebrate and invertebrate hosts might play important roles in the genetic variations of VP2 and VP7, respectively.
In recent years, epidemics caused by arboviruses have tended to spread over large areas due to the increasing activity of vector insects. Changes in the population of vector insects might be influencing the evolution of the viruses. Further study is needed to investigate the mutation of the virus.
Acknowledgments
This work was supported by grants received from the Ministry of Agriculture, Forestry, and Fishery of Japan.
REFERENCES
- 1.Bak, U. B., C. K. Cheong, H. I. Choi, C. W. Lee, and H. S. Oh. 1983. An outbreak of Ibaraki disease in Korea. Korean J. Vet. Res. 23:81-89. [Google Scholar]
- 2.Bonneau, K. R., N. Z. Zhang, W. C. Wilson, J. B. Zhu, F. Q. Zhang, Z. H. Li, K. L. Zhang, L. Xiao, W. B. Xiang, and N. J. MacLachlan. 2000. Phylogenetic analysis of the S7 gene does not segregate Chinese strains of bluetongue virus into a single topotype. Arch. Virol. 145:1163-1171. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, C. H., T. L. Barber, and M. M. Jochim. 1978. Antigenic relationship of Ibaraki, bluetongue, and epizootic hemorrhagic disease viruses. Vet. Microbiol. 3:15-22. [Google Scholar]
- 4.Cheney, I. W., M. Yamakawa, P. Roy, J. O. Mecham, and W. C. Wilson. 1996. Molecular characterization of the segment 2 gene of epizootic hemorrhagic disease virus serotype 2: gene sequence and genetic diversity. Virology 224:555-560. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, P. W., I. Sendow, E. Soleha, Sukarsih, N. T. Hunt, and S. Bahri. 1995. Australian-Indonesian collaboration in veterinary arbovirology-a review. Vet. Microbiol. 46:151-174. [DOI] [PubMed] [Google Scholar]
- 6.Della-Porta, A. J., A. R. Gould, B. T. Eaton, and D. A. McPhee. 1985. Biochemical characterization of Australian orbiviruses. Prog. Clin. Biol. Res. 178:337-345. [PubMed] [Google Scholar]
- 7.de Mattos, C. C., C. A. de Mattos, B. I. Osburn, M. Ianconescu, and R. Kaufman. 1991. Evidence of genome segment 5 reassortment in bluetongue virus field isolates. Am. J. Vet. Res. 52:1794-1798. [PubMed] [Google Scholar]
- 8.de Mattos, C. C., C. A. de Mattos, B. I. Osburn, and N. J. MacLachlan. 1994. Evolution of the L2 gene of strains of bluetongue virus serotype 10 isolated in California. Virology 201:173-177. [DOI] [PubMed] [Google Scholar]
- 9.de Mattos, C. C., C. A. de Mattos, N. J. MacLachlan, L. D. Giavedoni, T. Yilma, and B. I. Osburn. 1996. Phylogenetic comparison of the S3 gene of United States prototype strains of bluetongue virus with that of field isolates from California. J. Virol. 70:5735-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle, K. A. 1992. An overview and perspective on orbivirus disease prevalence and occurrence of vectors in Australia and Oceania, p.44-57. In T. L. Walton and B. I. Osburn (ed.), Bluetongue, African horse sickness, and related orbiviruses. CRC Press, Inc., Boca Raton, Fla.
- 11.Gould, A. R. 1987. The complete nucleotide sequence of bluetongue virus serotype 1 RNA3 and a comparison with other geographic serotypes from Australia, South Africa and the United States of America, and with other orbivirus isolates. Virus Res. 7:169-183. [DOI] [PubMed] [Google Scholar]
- 12.Gould, A. R., and L. I. Pritchard. 1991. Phylogenetic analyses of the complete nucleotide sequence of the capsid protein (VP3) of Australian epizootic haemorrhagic disease of deer virus (serotype 2) and cognate genes from other orbiviruses. Virus Res. 21:1-18. [DOI] [PubMed] [Google Scholar]
- 13.Hammami, S., and B. I. Osburn. 1992. Analysis of genetic variation of epizootic hemorrhagic disease virus and bluetongue virus field isolates by coelectrophoresis of their double-stranded RNA. Am. J. Vet. Res. 53:636-642. [PubMed] [Google Scholar]
- 14.Huismans, H., and B. J. Erasmus. 1981. Identification of the serotype-specific and group-specific antigens of bluetongue virus. Onderstepoort J. Vet. Res. 48:51-58. [PubMed] [Google Scholar]
- 15.Iwata, H., S. Manabe, A. Yoshida, E. M. Pereira, and T. Inoue. 2001. The complete nucleotide sequences of L3 and S7 segments of Ibaraki virus encoding for the major inner capsid proteins, VP3 and VP7. J. Vet. Med. Sci. 63:73-78. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Liao, Y. K., Y. S. Lu, S. T. Huang, and S. H. Lee. 1996. The etiological and epidemiological studies on the Ibaraki disease in Taiwan. Exp. Rep. Taiwan Anim. Health Res. Inst. 32:49-57. [Google Scholar]
- 18.Miura, Y., Y. Inaba, T. Tsuda, S. Tokuhisa, K. Sato, H. Akashi, and M. Matsumoto. 1982. A survey of antibodies to arthropod-borne viruses in Indonesian cattle. J. Vet. Med. Sci. 44:857-863. [DOI] [PubMed] [Google Scholar]
- 19.Nara Pereira, E. M., T. Nishida, R. Tokunaga, H. Iwata, and T. Inoue. 2000. Cloning and expression of the M5 RNA segment encoding outer capsid VP5 of epizootic hemorrhagic disease virus Japan serotype 2, Ibaraki virus. J. Vet. Med. Sci. 62:301-304. [DOI] [PubMed] [Google Scholar]
- 20.Nara Pereira, E. M., H. Iwata, and T. Inoue. 2000. The complete nucleotide sequence of segment L2 of Ibaraki virus encoding for the antigen recognized by neutralizing antibodies. J. Vet. Med. Sci. 62:317-321. [DOI] [PubMed] [Google Scholar]
- 21.Ohashi, S., K. Yoshida, Y. Watanabe, and T. Tsuda. 1999. Identification and PCR-restriction fragment length polymorphism analysis of a variant of the Ibaraki virus from naturally infected cattle and aborted fetuses in Japan. J. Clin. Microbiol. 37:3800-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omori, T., Y. Inaba, T. Morimoto, Y. Tanaka, R. Ishitani, H. Kurogi, K. Munakata, and M. Matumoto. 1969. Ibaraki virus, an agent of epizootic disease of cattle resembling bluetongue. I. Epidemiologic, clinical and pathologic observations and experimental transmission to calves. Japan J. Microbiol. 13:139-157. [DOI] [PubMed] [Google Scholar]
- 23.Omori, T., Y. Inaba, T. Morimoto, Y. Tanaka, M. Kono, H. Kurogi, and M. Matumoto. 1969. Ibaraki virus, an agent of epizootic disease of cattle resembling bluetongue. II. Isolation of the virus in bovine cell culture. Japan J. Microbiol. 13:159-168. [DOI] [PubMed] [Google Scholar]
- 24.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard, L. I., A. R. Gould, W. C. Wilson, L. Thompson, P. P. Mertens, and A. M. Wade-Evans. 1995. Complete nucleotide sequence of RNA segment 3 of bluetongue virus serotype 2 (Ona-A). Phylogenetic analyses reveal the probable origin and relationship with other orbiviruses. Virus Res. 35:247-261. [DOI] [PubMed] [Google Scholar]
- 26.Ramig, R. F., C. Garrison, D. Chen, and D. Bell-Robinson. 1989. Analysis of reassortment and superinfection during mixed infection of Vero cells with bluetongue virus serotypes 10 and 17. J. Gen. Virol. 70:2595-2603. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Siaz-Ruiz, J. R., and J. M. Kaper. 1978. Isolation of double strand RNAs using LiCl fractionation procedure. Prep. Biochem. 8:1-17. [DOI] [PubMed] [Google Scholar]
- 29.Sugiyama, M., N. Hirayama, H. Sasaki, T. Sugimura, N. Minamoto, and T. Kinjo. 1987. Antigenic relationship among strains of Ibaraki virus and epizootic haemorrhagic disease virus studied with monoclonal antibodies. Res. Vet. Sci. 46:283-285. [PubMed] [Google Scholar]
- 30.Tan, B. H., E. Nason, N. Staeuber, W. Jiang, K. Monastryrskaya, P. Roy. 2001. RGD tripeptide of bluetongue virus VP7 protein is responsible for core attachment to Culicoides cells. J. Virol. 75:3937-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, W. C., H. C. Ma, E. H. Venter, A. A. van Djik, B. S. Seal, and J. O. Mecham. 2000. Phylogenetic relationships of bluetongue viruses based on gene S7. Virus Res. 67:141-151. [DOI] [PubMed] [Google Scholar]
- 33.Xu, G., W. Wilson, J. Mecham, K. Murphy, E. M. Zhou, and W. Tabachnick. 1997. VP7: an attachment protein of bluetongue virus for cellular receptors in Culicoides variipennis. J. Gen. Virol. 78:1617-1623. [DOI] [PubMed] [Google Scholar]