Abstract
If parasite genotype influences the clinical course of malaria, we expect that isolates from patients with similar pathology would be more closely related than would be expected by chance. To explore this prediction, we typed nine microsatellite markers in sympatric Plasmodium falciparum isolates from cerebral and uncomplicated malaria patients from Vietnam. Temporal structure and linkage disequilibrium were also examined in this data set.
Most inhabitants in malarious areas sooner or later become infected with Plasmodium falciparum, but few of them experience life-threatening complications (9). Despite our increasing knowledge of the molecular basis of severe malaria (6), determinants of outcome of individual infections remain unknown. Both host and parasite genetics clearly play a role. Host genetics accounted for 7 to 10% of the variation in frequency and clinical outcome of P. falciparum infections in a pedigreed human population in Sri Lanka (18), and parasite genotype was shown to influence the course of induced P. falciparum infections in humans (14, 15) and squirrel monkeys (7).
The role of parasite genetics in malaria pathogenesis is harder to evaluate in natural infections. Mathematical modeling has been used to argue that cerebral malaria is caused by a minority of virulent parasite strains (10). The reported space-time clustering of severe malaria in Kenya (23) and the association between two P. falciparum surface antigen variants in isolates from severe malaria in French Guyana (4) are consistent with this hypothesis. If parasite genetics plays a major role in pathogenesis, we would expect closely related parasites to be associated with similar disease expression. Here, we assessed genetic the relatedness of isolates from cerebral and uncomplicated malaria patients to explore this prediction.
Between January 1996 and January 1997, we prospectively studied 101 patients, aged 3 to 77 years, presenting at Lam Dong Provincial Hospital No. 2 in the town of Bao Loc, Vietnam, with microscopically confirmed P. falciparum infection. Clinical details were recorded, and parasite isolates were stored for DNA extraction (8). Bao Loc is situated 150 km northeast of Ho Chi Minh City, in an area of mesoendemicity on the southern highlands of Vietnam (8). The population served by the hospital (140,000 people) includes ethnic Vietnamese and ethnic minorities who inhabit remote hill areas (13). Eight patients, aged 11 to 35 years, were admitted to the intensive care unit with strictly defined cerebral malaria (25); the remainder were outpatients with uncomplicated P. falciparum infection. We excluded patients with complications other than cerebral malaria, since they are unlikely to be associated with common P. falciparum virulence factors (17). All patients or guardians provided informed consent, and the research protocol was approved by the relevant ethical committees at all institutions involved in this study.
Nine single-copy microsatellite markers were typed (chromosome location in parentheses): Polyα (4), TA81 (5), TA1 and TA87 (6), 2490 (10), ARAII (11), PfG377 and PfPK2 (12), and TA60 (13). Alleles were amplified by seminested PCR by using primers corresponding to conserved regions flanking the microsatellites. Primer sequences and PCR conditions were exactly as described elsewhere (3). One of the internal primers used in the second round of amplification was end labeled with either of four different fluorescent markers, so that reactions could be multiplexed and allele length and peak heights could be measured in an automated sequencer. Length variation of PCR products was measured on an ABI3100 DNA analyzer (Applied Biosystems, Foster City, Calif.). We scored multiple alleles at a given locus if minor peaks were more than one-third the height of the predominant peak; infections were considered to contain multiple clones if one or more loci showed more than one allele (2). Samples were randomized on microtiter plates for amplification, to avoid systematic bias. Repeatability was assessed by reamplifying and rescoring the nine loci from 27 randomly selected isolates. Identical results were obtained for 96% of replicates; eight of nine discrepancies occurred in infections with multiple alleles.
Multiple alleles were found in 40 (39.6%) isolates, with a maximum of three alleles/isolate. Levels of P. falciparum genetic diversity in southern Vietnam (Table 1) were intermediate between those in South America and Africa (2). Diversity was also higher than those observed on the Thai-Burmese border: seven of nine loci showed higher expected heterozygosity (2). To avoid biases due to equivocal assignment of haplotypes in multiple-clone infections, we analyzed single-clone infections separately and obtained similar heterozygosity values (mean, 0.69). Proportions of alleles shared between haplotypes, a measure of genetic relatedness, were used to construct a neighbor-joining tree (22) using the PHYLIP, version 3.5c, package, distributed by its author (J. Felsenstein) at http://evolution.genetics.washington.edu. Nine of the 85 different haplotypes were recovered from more than one patient, and one haplotype was found in six isolates sampled in the first, second, and fourth trimesters of the study (Fig. 1).
TABLE 1.
Number of different alleles and expected heterozygosity at nine microsatellite loci and standardized indices of association of alleles in haplotypes in isolates of P. falciparum from southern Vietnama
| Locus | No. of alleles | Expected heterozygosity |
|---|---|---|
| Polyα | 18 | 0.91 |
| TA81 | 9 | 0.77 |
| TA1 | 9 | 0.69 |
| TA87 | 9 | 0.76 |
| 2490 | 2 | 0.47 |
| ARAII | 9 | 0.82 |
| PfG377 | 4 | 0.48 |
| PfPK2 | 8 | 0.80 |
| TA60 | 7 | 0.60 |
| Mean | 8.3 | 0.70 |
Expected heterozygosity, a measure of genetic diversity, was calculated as [n/(n − 1)][1 − Σpi2], where n is the number of isolates sampled and pi is the frequency of each allele at a given locus (2). Allele frequencies were measured using only the predominant alleles present at each locus (i.e., that giving the highest peak) in cases of multiple-clone infection. The standardized index of association was calculated as VD/(Ve − 1)(r − 1), where VD is the observed variance, Ve is the expected variance under the hypothesis of random association of alleles in haplotypes, and r is the number of loci analyzed (11). Values were 0.021 (P < 0.0001), 0.033 (P < 0.0001), and 0.001 (P = 0.3930) for all infections, single-clone infections, and unique haplotypes, respectively. Single-clone infections (n = 61) were analyzed separately to detect possible biases due to equivocal assignment of haplotypes in multiple-clone infections. Unique haplotypes (n = 85) were analyzed separately to distinguish between clonal and “epidemic” population structure of parasites (19).
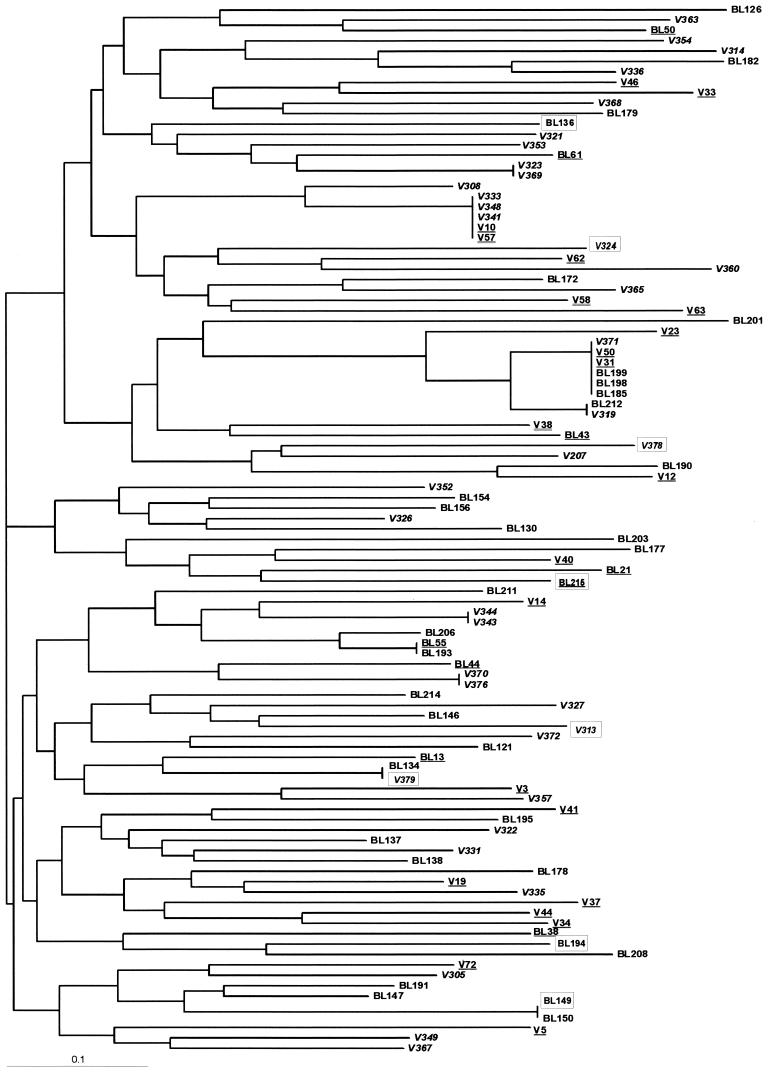
FIG. 1.
Neighbor-joining tree showing the relationships between microsatellite haplotypes of 101 P. falciparum isolates from southern Vietnam. The predominant allele at each locus was used to define nine-locus parasite haplotypes in multiple-clone infections. Dates of collection of isolates are indicated as follows: double underlining (n = 22), January to April 1996; italics (n = 37), May to July 1996; single underlining (n = 8), August to October 1996; and normal type (n = 34), November 1996 to January 1997. Terminal branches of zero length mark identical haplotypes. Isolates from cerebral malaria patients (n = 8) are boxed.
Visual inspection of Fig. 1 revealed no apparent temporal clustering of haplotypes. We found a weak correlation between genetic distance and time (i.e., interval between dates of collection) by using a Mantel nonparametric test (19) (r = 0.038, P = 0.07). The weak evidence for temporal clustering in Vietnam contrasts dramatically with that observed in Venezuela (24), probably due to differences in levels of malaria transmission and effective parasite population sizes in these two regions.
The population structure of local isolates was assessed as described previously (2). LIAN 3.1 (11) was used to compute the proportion of alleles shared in pairwise comparisons, to measure its variance and to compare this value with the variance expected under the hypothesis of random assortment of alleles in haplotypes. The strength of multilocus linkage disequilibrium was estimated with a standardized index of association (12). The highly significant deviation from random association between alleles disappeared when each haplotype was treated as an individual (20) (Table 1). These results provided evidence for an “epidemic” population structure, as defined by Maynard Smith and colleagues (20), characterized by linkage disequilibrium resulting from the propagation of a few haplotypes in an otherwise panmictic population. Similar results have been obtained for parasites from South America, Thailand, and Papua New Guinea (2).
Haplotypes from cerebral malaria patients did not cluster together in the neighbor-joining tree, and two pairs of identical haplotypes were recovered from patients with discordant clinical outcomes, either cerebral or uncomplicated malaria (Fig. 1). Mean genetic distances (1 − proportion of shared alleles) were compared between haplotypes recovered from cerebral malaria patients (0.754; 28 pairwise comparisons), between haplotypes from uncomplicated malaria patients (0.716; 4,278 pairwise comparisons), and between haplotypes from both groups of patients (0.698; 744 pairwise comparisons). Parasites from cerebral malaria cases therefore shared fewer alleles and were less closely related than parasites from uncomplicated malaria cases, in contrast to our expectations. However, randomization tests indicated that these differences were not significant. In conclusion, we found no evidence suggesting that cerebral malaria is caused by a subset of genetically related parasites in this population of patients. Interestingly, recent comparisons of antibody recognition of variant surface antigens (VSA) expressed by P. falciparum isolates from African children with severe and uncomplicated malaria also argue against the notion that more intense morbidity is associated with infections with relatively rare and virulent parasite variants, since common VSA (i.e., frequently recognized by antibodies of malaria-exposed people living in the same area) were predominantly expressed by parasites found in severe disease (5).
We used a set of nine putatively neutral microsatellite markers, instead of surface antigen genes under strong selection pressure (1, 4, 16, 21), to compare the genetic backgrounds of sympatric isolates from patients with cerebral and uncomplicated malaria. We tested the hypothesis that parasites causing cerebral malaria were more genetically related than expected by chance, instead of looking for associations between particular genotypes or antigenic variants and morbidity. Why did we find no evidence that parasite genotype influences malaria pathology in this data set? We suggest three possible reasons. (i) This and previous studies are underpowered due to the small number of isolates analyzed and limited resolution of parasite haplotypes. (ii) Interactions between host and parasite genotypes, rather than parasite genes on their own, may be critical in determining disease expression. (iii) If many of the parasites sampled are unrelated, this reduces the power of our approach. Frequent recombination breaks down associations between loci, and the relatedness between parasites is obscured. We note that a strong association between two surface antigen markers and pathology was recently found in a parasite population with much lower levels of recombination than those seen in Vietnam (4). Such parasite populations, in which cross-fertilization is extremely rare, may be optimal for disclosing associations between disease pathogenesis and parasite genotype. In contrast, no association between expression of particular (and relatively rare) VSA and severe malaria could be detected in an area of intense malaria transmission in Kenya, where recombination is very frequent (5).
Acknowledgments
This study was supported by grants from NIH (RO1 AI48071), Fundação de Amparo à Pesquisa do Estado de São Paulo (98/14587-1), and Toyota Foundation (96B3-001).
REFERENCES
- 1.Al-Yaman, F., B. Genton, J. C. Reeder, D. Mokela, R. F. Anders, and M. P. Alpers. 1997. Humoral response to defined Plasmodium falciparum antigens in cerebral and uncomplicated malaria and their relationship to parasite genotype. Am. J. Trop. Med. Hyg. 56:430-435. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, T. J. C., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. P. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, T. J. C., X. Z. Su, M. Bockerie, M. Lagog, and K. P. Day. 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119:113-126. [DOI] [PubMed] [Google Scholar]
- 4.Ariey, F., D. Hommel, C. Le Scanf, J. B. Duichemin, C. Peneau, A. Hulin, J. L. Sarthou, J. M. Reynes, T. Fandeur, and O. Mercereau-Puijalon. 2001. Association of severe malaria with a specific Plasmodium falciparum genotype in French Guyana. J. Infect. Dis. 184:237-241. [DOI] [PubMed] [Google Scholar]
- 5.Bull, P. C., M. Kortok, O. Kai, F. Ndungu, A. Ross, B. S. Lowe, C. I. Newbold, and K. Marsh. 2000. Plasmodium falciparum-infected erythrocytes: agglutination by diverse Kenyan plasma is associated with severe disease and young host age. J. Infect. Dis. 182:252-259. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Q., M. Schlichtherle, and M. Wahlgren. 2000. Molecular aspects of severe malaria. Clin. Microbiol. Rev. 13:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fandeur, T., O. Mercereau-Puijalon, and B. Bonnemains. 1996. Plasmodium falciparum: genetic diversity of several strains infectious for the squirrel monkey (Saimiri sciureus). Exp. Parasitol. 84:1-15. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira, M. U., Q. Liu, M. Zhou, M. Kimura, O. Kaneko, H. V. Thien, S. Isomura, K. Tanabe, and F. Kawamoto. 1998. Stable patterns of allelic diversity at the Merozoite Surface Protein-1 locus of Plasmodium falciparum in clinical isolates from southern Vietnam. J. Eukaryot. Microbiol. 45:131-136. [DOI] [PubMed] [Google Scholar]
- 9.Greenwood, B., K. Marsh, and R. W. Snow. 1991. Why do some children develop severe malaria? Parasitol. Today 7:277-281. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, S., A. S. V. Hill, D. Kwiatkowski, A. M. Greenwood, B. M. Greenwood, and K. P. Day. 1994. Parasite virulence and disease patterns in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 91:3715-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 12.Hudson, R. R. 1994. Analytical results concerning linkage disequilibrium in models with genetic transformation and recombination. J. E vol. Biol. 7:535-548. [Google Scholar]
- 13.Hyunh, T. V., C. V. Thanh, and A. T. Kim. 1990. Severe malaria in a provincial hospital in Vietnam. Lancet 336:1316.. [PubMed] [Google Scholar]
- 14.James, S. P., W. D. Nicol, and P. G. Shute. 1932. A study of induced malignant tertian malaria. Proc. R. Soc. Med. 25:1153-1181. [PMC free article] [PubMed] [Google Scholar]
- 15.James, S. P., W. D. Nicol, and P. G. Shute. 1936. Clinical and parasitological observations on induced malaria. Proc. R. Soc. Lond. B 29:879-894. [PMC free article] [PubMed] [Google Scholar]
- 16.Kun, J. F. J., R. J. Schmidt-Ott, L. G. Lehman, B. Lell, D. Luckner, B. Greve, P. Matousek, and P. G. Kremsner. 1998. Merozoite surface antigen 1 and 2 genotypes and rosetting of Plasmodium falciparum in severe and mild malaria in Lambaréné, Gabon. Trans. R. Soc. Trop. Med. Hyg. 92:110-114. [DOI] [PubMed] [Google Scholar]
- 17.Kurtzhals, J. A. L., B. Q. Goka, B. D. Akanmori, and L. Hviid. 2001. The importance of strict patient definitions in studies of malaria pathogenesis. Parasitol. Today 17:313-314. [DOI] [PubMed] [Google Scholar]
- 18.Mackinnon, M. J., D. M. Gunawardena, J. Rajakaruna, S. Weerasingha, K. N. Mendis, and R. Carter. 2000. Quantifying genetic and nongenetic contributions to malarial infection in a Sri Lanka population. Proc. Natl. Acad. Sci. USA 97:12661-12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 20.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert, F., F. Ntoumi, G. Angel, D. Candito, C. Rogier, T. Fandeur, J. L. Sarthou, and O. Mercereau-Puijalon. 1996. Extensive genetic diversity of Plasmodium falciparum isolates collected from patients with severe malaria in Dakar, Senegal. Trans. R. Soc. Trop. Med. Hyg. 90:704-711. [DOI] [PubMed] [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Snow, R. W., J. R. Schellenberg, N. Peshu, D. Foster, C. R. Newton, P. A. Winstanley, I. Mwangi, C. Waruiru, P. A. Warn, C. Newbold, and K. Marsh. 1993. Periodicity and space-time clustering of severe childhood malaria on the coast of Kenya. Trans. R. Soc. Trop. Med. Hyg. 87:386-390. [DOI] [PubMed] [Google Scholar]
- 24.Urdaneta, L., A. Lal, C. Barnabé, B. Oury, I. Goldman, F. J. Ayala, and M. Tibayrenc. 2001. Evidence for clonal propagation in natural isolates of Plasmodium falciparum from Venezuela. Proc. Natl. Acad. Sci. USA 98:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warrell, D. A., M. Molyneaux, and P. F. Beasles. 1990. Severe and complicated malaria. Trans. R. Soc. Trop. Med. Hyg. 84(Suppl. 2):1-65. [Google Scholar]