Abstract
Multilocus sequence typing (MLST) based on the 16S RNA, pduF, glnA, and manB genes was developed for Salmonella, and its discriminatory ability was compared to those of pulsed-field gel electrophoresis (PFGE) and serotyping. PFGE differentiated several strains undifferentiable by serotyping, and 78 distinct PFGE types were identified among 231 Salmonella isolates grouped into 22 serotypes and 12 strains of undetermined serotype. The strains of several PFGE types were further differentiated by MLST, which suggests that the discriminatory ability of MLST for the typing of Salmonella is better than that of serotyping and/or PFGE typing. manB-based sequence typing identified two distinct genetic clusters containing 32 of 54 (59%) clinical isolates whose manB gene sequences were analyzed. The G+C contents and Splitstree analysis of the manB, glnA, and pduF genes of Salmonella indicated that the genes differ in their evolutionary origins and that recombination played a significant role in their evolution.
Nontyphoidal salmonellae are among the leading causes of food-borne disease in the United States, in which they cause approximately 1.4 million cases of salmonellosis each year (29). In 50 to 75% of the cases, the etiological agent is acquired from meat, poultry, or eggs, with poultry considered the primary vehicle of transmission (18). Because of the importance of Salmonella in food-borne disease, numerous typing methodologies have been developed and have been used to trace salmonellosis outbreaks to the contaminated source and to delineate the epidemiology of Salmonella infections. Serotyping of the phase 1 and 2 flagellar proteins (the H1 and H2 antigens, respectively) and the O-specific polysaccharide (O antigen) in the bacterium's lipopolysaccharide-containing outer membrane is one of the most commonly used approaches to the characterization of Salmonella strains (5). However, the genes encoding O antigens and phase I flagellin have been shown (27, 31) to be extremely variable and highly prone to recombination, which has so far resulted in the identification of more than 2,400 distinct serotypes of Salmonella. This abundance of serotypes, together with the fact that some serotypes cross-react with Escherichia coli and other bacterial species and that special adsorbed sera (not readily available in most clinical and research laboratories) are required for serotyping, limits the usefulness of the assay. Other phenotypic approaches, including phage typing, biotyping, and antibiotic susceptibility testing (20, 36), also have been used for the characterization of Salmonella strains. However, these techniques are tedious and have other important drawbacks (e.g., phage typing requires specific typing phages, and its reproducibility is of concern). Most importantly, the discriminatory abilities of these approaches are not optimal, and they frequently fail to discriminate between epidemiologically related and unrelated Salmonella strains (6, 40).
The recent introduction of modern phenotype-based methodologies for the typing of Salmonella strains, such as multilocus enzyme electrophoresis (2), as well as DNA-based methodologies, including arbitrarily primed PCR (15), pulsed-field gel electrophoresis (PFGE) (32), and ribotyping (39), has addressed some of these problems. However, these methods, although generally superior to serotyping and phage typing, also vary in their reproducibilities and discriminatory abilities. PFGE is recognized (3, 4, 32) to be superior to the other methods and therefore is considered the “gold standard” for the subtyping of Salmonella strains. However, the discriminatory ability of PFGE also is not optimal, and despite strenuous efforts at standardization, there may be striking variability in PFGE gels (and data interpretation) among various laboratories. Thus, approaches which are more discriminatory and less prone to human error are required for epidemiologic investigations of Salmonella outbreaks. In this context, a recently developed (28) methodology called multilocus sequence typing (MLST) may provide an ideal balance of high discriminatory power and a powerful data analysis capability requiring minimal human input. This technique, made possible by the increased availability of robotic sequencers, is based on determination of the nucleotide sequences of a series of predetermined housekeeping, ribosomal, and/or virulence-associated genes. Thus, MLST provides data similar to those obtained by multilocus enzyme electrophoresis, but in substantively greater detail, because it has the ability to assess individual nucleotide changes rather than to screen for changes in the overall charge and expression of the enzyme under study (28). MLST has rapidly been gaining increased recognition as one of the best molecular typing approaches available today, and it has been used to characterize several pathogenic bacteria, including Neisseria meningitidis (16), Staphylococcus aureus (12), Yersinia pestis (1), Streptococcus pneumoniae (11, 17), Vibrio cholerae (35), and, most recently, Campylobacter jejuni (9). However, information about MLST of Salmonella is not available in the peer-reviewed literature, and this is the first report describing the development and use of MLST to characterize this important human pathogen.
The initial aims of this study were to develop the MLST approach for Salmonella (as a possible tool for epidemiologic investigations of salmonellosis outbreaks) and to compare the discriminatory ability of MLST with those of PFGE and serotyping. However, the MLST data generated during the course of our studies also provided us with new insights into the clonal relatedness between environmental and clinical Salmonella isolates, as well as with an increased appreciation of the role of recombination in the genetic diversity of Salmonella strains.
MATERIALS AND METHODS
Bacterial strains.
A total of 243 Salmonella strains (182 environmental isolates [obtained primarily from poultry and poultry farms] and 61 clinical isolates) isolated in Maryland from 1996 to 1999 were characterized in this study. In this collection, 231 strains were grouped into 22 serotypes, and the remaining 12 strains were of unknown serotype (phage-typing information was not available for the strains) (Table 1). The standard strain Salmonella enterica subsp. enterica serotype Newport 01144 from the Centers for Disease Control and Prevention (CDC) was included in the strain collection mentioned above, and it was used as a reference strain during typing by PFGE.
TABLE 1.
Salmonella strains used in the study by serotype and source of isolation, PFGE types and the number of strains by PFGE type, and the number of strains analyzed by MLST of each of the four genes
| Serotype | No. of strains (cl/env)a | Strains tested, by PFGE type | No. of strains analyzed for the following genes by MLST:
|
|||
|---|---|---|---|---|---|---|
| manB | glnA | pduF | 16S RNA | |||
| Hadar | 83 (0/83) | Type P5, 19, 29, 30, 48, 49, 50, 63, 71, 74, 77, 78, 171, 173, and 182; type P40, 82; type P41, 83; type P53, 120; type P55, 122; type P67, 143, 144, 149, 172, 175, 176, 177, and 183; type P68, 55, 56, 58, 60, 102, 104, 118, 119, 124, 125, 126, 127, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 145, 146, 147, 148, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 174, 179, 180, 181, 184, 185, and 186 | 79 | 80 | 64 | 79 |
| Typhimurium | 42 (15/27) | Type P2, 6 and 59; type P3, 7, 14, 15, 31, 32, 51-1, 52-1, 54-1, 55-1, 13-1, 14-1, 15-1, 16-1, 18-1, 11-1, 12-1, 20-1; type P4, 2, 13, 84, 85, 86, 87, 88, 89, 90, 91, 92, 107, and 53-1; type P19, 19-1; type P21; 46, 54, and 61; type P22, 47 and 62; type P23, 51; type P24, 52; type P47, 108; type P57, 17-1 | 41 | 39 | 30 | 39 |
| Enteritidis | 24 (15/9) | Type P1, 1, 5, 21, 24, 34, 98, 106, 24-1, 25-1, 26-1, 27-1, 28-1, 29-1, 30-1, 56-1, 57-1, 58-1, 21-1, and 22-1; type P6, 4; type P7, 33; type P76, 59-1; type P60, 60-1; type P59, 23-1 | 24 | 24 | 17 | 22 |
| Heidelberg | 21 (15/6) | Type P9, 9, 57, and 67; type P10, 12; type P11, 35; type P50, 113, 1-1, 2-1, 3-1, 4-1, 5-1, 6-1, 7-1, 8-1, 9-1, 46-1, 47-1, and 50-1; type P74, 49-1; type P69, 10-1; type P73, 48-1 | 17 | 18 | 12 | 17 |
| Newportb | 19 (16/3) | Type P26, 26, 36-1, and 38-1; type P37, 76; type P48, 001144; type P60, 31-1, 33-1, and 45-1; type P61, 32-1; type P62, 34-1; type P63, 35-1; type P64, 37-1; type P65, 39-1; type P66, 40-1; type P70, 41-1; type P71, 42-1, and 43-1; type P72, 44-1; type P77, 128 | 15 | 14 | 13 | 15 |
| Kentuckyc | 9 (0/9) | Non typeable by PFGE, 10, 11, 23, 38, 39, 66, 69, 103, 105 | 9 | 7 | 7 | 4 |
| Agona | 5 (0/5) | Type P20, 36; type P36, 75; type P52, 117; type P54, 121; type P78, 123 | 6 | 5 | 5 | 6 |
| Reading | 5 (0/5) | Type P49, 111, 112, 114, 115, and 116 | 4 | 1 | 5 | 5 |
| Senftenberg | 4 (0/4) | Type P18, 18; type P32, 28; type P43, 94; type P44, 95 | 4 | 4 | 4 | 4 |
| Mbandaka | 3 (0/3) | Type P17, 17; type P29, 22; type P31, 27 | 3 | 3 | 2 | 3 |
| Schwarzengrund | 2 (0/2) | Type P8, 40 and 42 | 2 | 2 | 2 | 2 |
| Cambridge | 2 (0/2) | Type P42; 93 and 97 | 2 | 2 | 2 | 2 |
| Johannesburg | 2 (0/2) | Type P56, 99 and 100 | 2 | 2 | 2 | 1 |
| Alachua | 1 (0/1) | Type P13, 16 | 1 | 1 | 1 | 1 |
| Infantis | 1 (0/1) | Type P15, 43 | 1 | 1 | 0 | 1 |
| Tennessee | 1 (0/1) | Type P14, 41 | 1 | 1 | 1 | 1 |
| Worthington | 1 (0/1) | Type P16, 44 | 1 | 1 | 1 | 1 |
| Haardt | 1 (0/1) | Type P19, 20 | 1 | 1 | 1 | 1 |
| Indiana | 1 (0/1) | Type P30, 25 | 1 | 1 | 0 | 1 |
| Brandenberg | 1 (0/1) | Type P34, 68 | 1 | 1 | 1 | 1 |
| Anatum | 2 (0/2) | Type P12, 37; type P45, 95 | 2 | 2 | 2 | 0 |
| Saint paul | 1 (0/1) | Type P33, 70 | 1 | 1 | 0 | 1 |
| Undeterminedc | 12 (0/12) | Type P25, 53; type P27, 3; type P28, 8; type P35, 45 and 109; type P38, 79; type P39, 80 and 81; type P46, 101; type P51, 110; nontypeable by PFGE; 64 and 65 | 11 | 12 | 11 | 10 |
| Total | 243 (61/182) | 229 | 223 | 183 | 217 | |
cl/env, number of clinical isolates/number of environmental isolates (all clinical strains are hyphenated; e.g., strain 52-1 is a clinical isolate, and strain 52 is an environmental isolate).\
Includes CDC standard strain Salmonella serotype Newport 01144.\
The serotype Kentucky strains and two strains of undetermined serotypes could not be typed by the standard Salmonella PFGE-typing protocol.
PFGE.
PFGE was performed as described previously (19) with a CHEF DR II apparatus (Bio-Rad Laboratories, Hercules, Calif.). The DNA in the plugs was digested by incubation (37°C, 4 h) of the plugs with XbaI, and electrophoresis was performed in a 1% agarose gel (in 0.5× TBE [Tris-borate-EDTA] buffer). The following electrophoresis conditions were used: voltage, 180; initial time, 2.2 s; final time, 64 s; run time, 20 h. Bacteriophage lambda ladder pulsed-field grade (PFG) and low-range PFG molecular weight markers were loaded onto all gels. XbaI-digested DNA from Salmonella serotype Newport strain 01144 was used as the reference in all experiments.
MLST.
The four Salmonella loci selected for MLST analysis were the genes encoding 16S RNA, phosphomannomutase (manB), glutamine synthetase (glnA), and the 1,2-propanediol utilization factor (pduF). The primers were designed by analyzing corresponding Salmonella and E. coli gene sequences in GenBank (http://www.ncbi.nlm.nih.gov). The sequences were aligned by use of ClustalX (24), and primers were selected from conserved regions flanking potentially variable internal fragments of the targeted genes (Table 2). The same primers were used for PCR amplification and sequencing.
TABLE 2.
Primers used for MLST of a subset of 107 Salmonella strains, the number of alleles and polymorphic sites identified per gene, and the dN/dS ratios for various genes
| Gene | Primers (5′ → 3′) | Size (bp) of fragment analyzed | No. of alleles | No. of polymorphic sites | Proportion (%) of polymorphic sites | Mean G+C content (%) | dN/dS |
|---|---|---|---|---|---|---|---|
| 16S rRNA | AGTTTGATCATGGCTCAG | 350 | 9 | 2 | 0.6 | 56 | NAa |
| TTACCGCGGCTGGCA | |||||||
| manB | CCGGCACCGAAGAGA | 660 | 32 | 37 | 5.6 | 63 | 0.1870 |
| CGCCGCCATCCGGTC | |||||||
| pduF | CT(C/A)AAAGTCGCYGGYGC | 210 | 17 | 7 | 3.3 | 51 | 0.3101 |
| GGGTTCATTGCAAAACC | |||||||
| gluA | CCGCGACCTTTATGCCAAAACCG | 270 | 15 | 9 | 3.3 | 61 | 0.0807 |
| CCTGTGGGATCTCTTTCGCT |
NA, not applicable.
Bacterial DNA was extracted from the plugs prepared for PFGE. Briefly, plugs containing bacterial DNA were frozen and thawed (at −70°C and 55°C, respectively) twice in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), the supernatants were collected after clarification of the samples by centrifugation (5,000 × g, 10 min), and aliquots (1 μl) of the resulting supernatants (containing approximately 50 ng of bacterial DNA) were used as templates for PCR amplification. For most samples, the PCR amplification conditions were 94°C for 5 min, followed by 35 amplification cycles, each consisting of sequential incubation at 94°C (45 s), 55°C (45 s), and 72°C (5 min). When the primers for the glnA gene were used, a reduced annealing temperature (44°C instead of 55°C) was sometimes required in order to achieve reproducible amplification. Sequencing of amplified fragments was performed in both directions with a BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Inc., Foster City, Calif.). The labeled fragments were separated by size by using either an ABI 377 Prism automated sequencer or an ABI 3700 DNA analyzer (Applied Biosystems, Inc.).
Data analysis.
During PFGE analysis, the Salmonella isolates were separated according to their PFGE patterns, based on two band differences (38). The PFGE patterns were compared by means of the Dice coefficient with Fingerprinting DST Molecular Analyst software (Bio-Rad), and clustering of strains was based on the unweighted pair group method with averages (a tolerance of 3% in the band position was applied). The computer-assisted analysis was performed according to the instructions of the manufacturer.
For the analysis by MLST, the internal fragment sequences of the 16S rRNA, manB, glnA, and pduF genes from the subset of Salmonella isolates were analyzed (the number of strains analyzed varied for each locus; Table 1). The reading of trace files and the assembly of contigs (for each gene of each strain) were performed with the programs Phred (13, 14) and Phrap (available at http://www.washington.edu), respectively. The sequences were trimmed and aligned by use of the ClustalX program (24), and the genetic relatedness of the strains was estimated by the maximum parsimony and maximum likelihood approaches. Dendrograms for each of the four genes were constructed by the use of neighbor-joining and bootstrapping algorithms, as implemented in PAUP (37). The sequence type analysis and recombinational tests (START) program (http://outbreak.ceid.ox.ac.uk) was used to determine the G+C content, the number of alleles, the proportion of polymorphic sites, and the proportion of nonsynonymous and synonymous base substitutions (dN and dS, respectively). The bootstrapping procedure for splits decomposition analysis, as implemented in the Splitstree program (23), was used to test for parallel changes in the DNA sequences.
Nucleotide sequence accession numbers.
The DNA sequences of the 16S RNA, pduF, glnA, and glpF genes have been deposited in GenBank under accession numbers AF415245 through AF416102.
RESULTS
PFGE analysis.
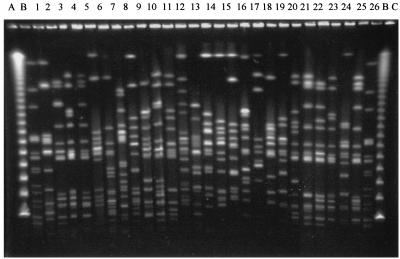
Seventy-eight distinct PFGE types were identified among the 232 strains (including a distinct PFGE type [type P48] for CDC standard strain 01144). Eleven strains (all nine Salmonella serotype Kentucky isolates and two strains of undetermined serotype) could not be typed by the standard Salmonella PFGE-typing procedure. Twenty-six representative PFGE patterns are presented in Fig. 1. Among the five serotypes represented by more than 15 strains (Table 1), serotype Newport contained the most PFGE types (1 PFGE type per 1.4 strains), followed by serotype Heidelberg (1 PFGE type per 3 strains), serotype Enteritidis (1 PFGE type per 4 strains), and serotype Typhimurium (1 PFGE type per 4.2 strains). The dendrogram constructed on the basis of the XbaI PFGE patterns (Fig. 2) revealed that the Salmonella serotype Newport strains were more scattered on the dendrogram than the other strains, e.g., serotype Enteritidis and Typhimurium strains, which formed relatively tighter clusters. PFGE differentiated strains within a single serotype (e.g., 10 PFGE types were identified within the Typhimurium serotype, and 7 PFGE types were identified within the Heidelberg serotype), but not vice versa; i.e., serotyping did not differentiate strains within a single PFGE type.
FIG. 1.
PFGE patterns of XbaI-digested DNA of Salmonella strains. Lane A, bacteriophage λ ladder PFG marker; lane B, CDC standard strain Salmonella serotype Newport 01144 (XbaI digest); lane C, low-range PFG marker; lanes 1 through 26, representative Salmonella PFGE types P1 through P26, respectively.
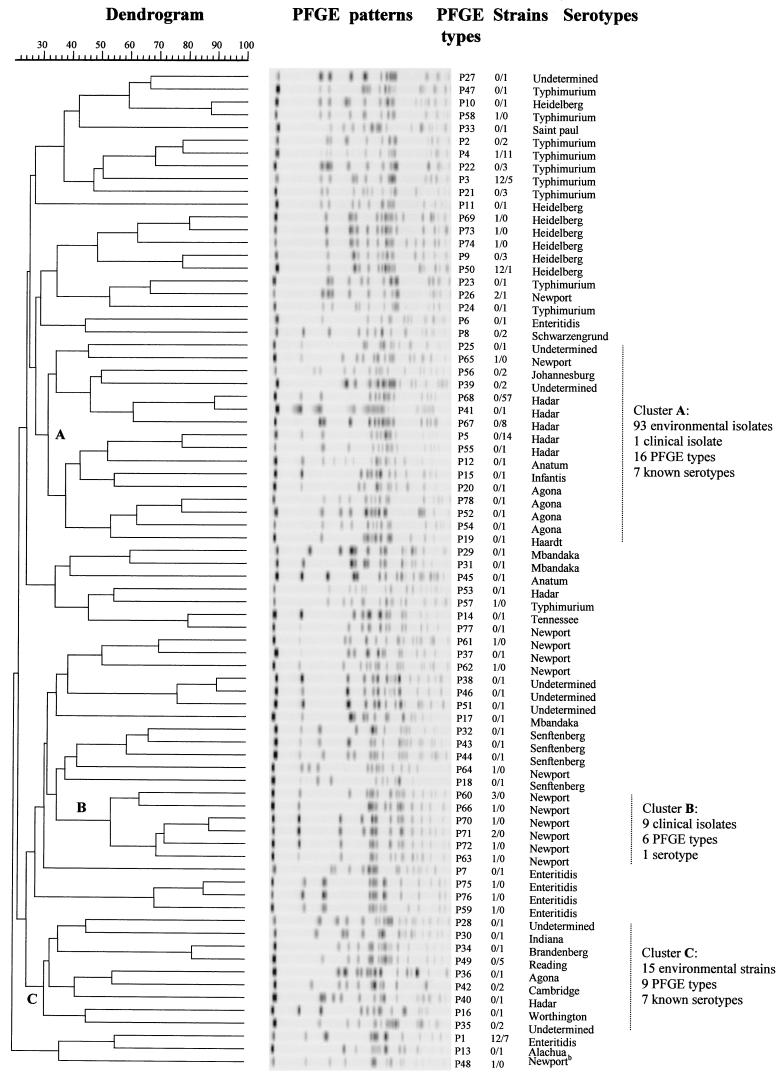
FIG. 2.
Dendrogram portraying the genetic diversity of various Salmonella strains, constructed on the basis of the PFGE patterns of XbaI-digested Salmonella DNA. The values in the Strains column indicate the number of clinical isolates/number of environmental isolates.
In order to determine whether genetically related strains clustered according to their source (clinical versus environmental), we defined the minimum size of a cluster as five closely related PFGE types, and we identified, on the basis of the dendrogram shown in Fig. 2, one cluster containing only clinical isolates (cluster B, encompassing six PFGE types and containing nine strains of Salmonella serotype Newport). In addition, we identified two large environmental clusters, clusters A and C, each of which contained 7 known serotypes and which contained 16 and 9 PFGE types, respectively, and 93 and 15 environmental isolates, respectively (cluster A also contained a single clinical isolate) (Fig. 2). The two environmental clusters primarily consisted of serotype Hadar isolates (82 [76%] of the 108 isolates in the environmental clusters belonged to serotype Hadar), but they also contained strains of several other serotypes (including serotype Newport) and 6 strains of undetermined serotype.
Analysis by MLST.
The number of isolates subjected to MLST varied for each locus and ranged from 183 strains analyzed for pduF gene sequences to 229 strains analyzed for manB gene sequences (Table 1). This variability in the number of strains for each of the four genes was caused by differences in the number of strains whose fragments were amplified during the PCR (e.g., the primers specific for the 16S rRNA, manB, glnA, and pduF genes amplified those genes from 89, 94, 92, and 75% of the strains in our strain collection, respectively), and exhaustive attempts were not made to analyze the same number of strains for each of the four loci.
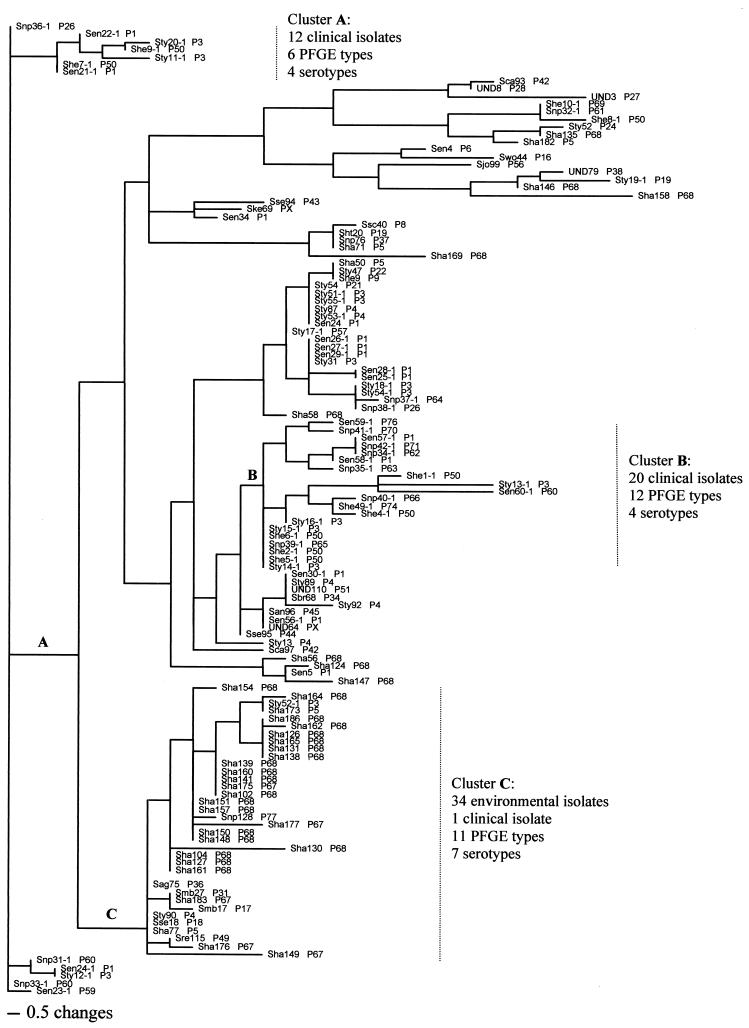
The DNA sequences of each of the four genes were analyzed by the maximum parsimony and maximum likelihood methods, previously reported (21) to be well suited for determination of phylogenetic relationships among various bacterial strains and species, and both methods gave consistent results for each of the four genes. Clustering of clinical versus environmental isolates was not observed in analyses with the 16S RNA, pduF, and glnA gene fragments. In contrast, clustering of clinical isolates did occur when the manB gene fragments were analyzed, and the dendrogram (Fig. 3) revealed two distinct clusters containing only clinical isolates (clusters A and B, each of which contained four serotypes and which contained 12 and 20 strains, respectively, and 6 and 12 PFGE types, respectively). Another large cluster (cluster C) contained 34 environmental strains of 11 PFGE types and 7 serotypes and a single clinical isolate (Fig. 3). The clusters containing clinical isolates (clusters A and B) encompassed 32 of the 54 (ca. 59%) clinical isolates, and the cluster containing environmental isolates (cluster C) encompassed 34 of the 175 (ca. 19%) environmental isolates whose manB gene sequences were analyzed. As with the other three genes, several clusters containing both environmental and clinical isolates were also identified.
FIG. 3.
Neighbor-joining tree of Salmonella isolates, constructed by the maximum parsimony method by using the sequences of the manB gene fragments. The designations at the branches indicate the following: serotype, strain number (all hyphenated strains are clinical isolates), and PFGE type (PX, strains not typeable by PFGE). The following serotype designations are used: Sag, serotype Agona; San, serotype Anatum; Sbr, serotype Branderberg; Sca, serotype Cambridge; Sen, serotype Enteritidis; Sht, serotype Haardt; Sha, serotype Hadar; She, serotype Heidelberg; Sjo, serotype Johannesburg; Ske, serotype Kentucky; Smb, serotype Mbandaka; Snp, serotype Newport; Sre, serotype Reading; Sse, serotype Senftenberg; Sty, serotype Typhimurium; Ssc, serotype Schwarzengrund; Swo, serotype Worthington; and UND, undetermined serotype. For example, the branch labeled Snp36-1 P26, at the top of the figure indicates Salmonella serotype Newport, strain 36-1, PFGE type P26. Horizontal length represents genetic distance, and vertical lengths are not meaningful. For the sake of space, the figure contains a short version of the dendrogram, containing 127 strains from among a total of 229 isolates whose manB gene sequences were analyzed.
Correlation between PFGE and MLST.
Several strains within the same PFGE type were clustered into separate types by MLST, but not vice versa (i.e., sequence-based clusters were not confined to a single PFGE type or cluster). For example, Salmonella serotype Hadar strains 104, 135, 164, and 169 (and some other serotype Hadar isolates) were clustered into a single PFGE type by PFGE typing (type P68); however, they were separated into distinct (and often not closely related) sequence types on the basis of sequence typing of their manB loci (Fig. 3) and their glnA and pduF loci (data not shown). As expected, the 16S RNA-encoding gene was highly conserved and grouped most isolates together (data not shown).
Allele and polymorphic site distributions.
In order to determine the allele distribution and the number of polymorphic sites (and their proportions) in the four gene fragments analyzed, we used the START program to examine a subset of 107 strains (18 clinical isolates and 89 environmental isolates) for which all sequences were available. The number of alleles varied for the four genes and was the smallest for the 16S RNA fragments, in which only nine alleles were identified. The numbers of alleles were 15, 17, and 32 for the glnA, pduF, and manB fragments, respectively (Table 2). The proportions of polymorphic sites also varied for the four genes, ranging from 0.6% for the 16S RNA fragments to 5.6% for the manB locus. The proportion of nucleotide substitutions that changed the amino acid sequence (nonsynonymous base substitutions [dN]) and the proportion that did not (synonymous base substitutions [dS]) were calculated, and the dN/dS ratios were determined to be less than 1 for the manB, pduF, and glnA genes (Table 2). A similar analysis could not be performed with the 16S RNA sequences because they do not contain open reading frames.
Splitstree analysis for parallel changes.
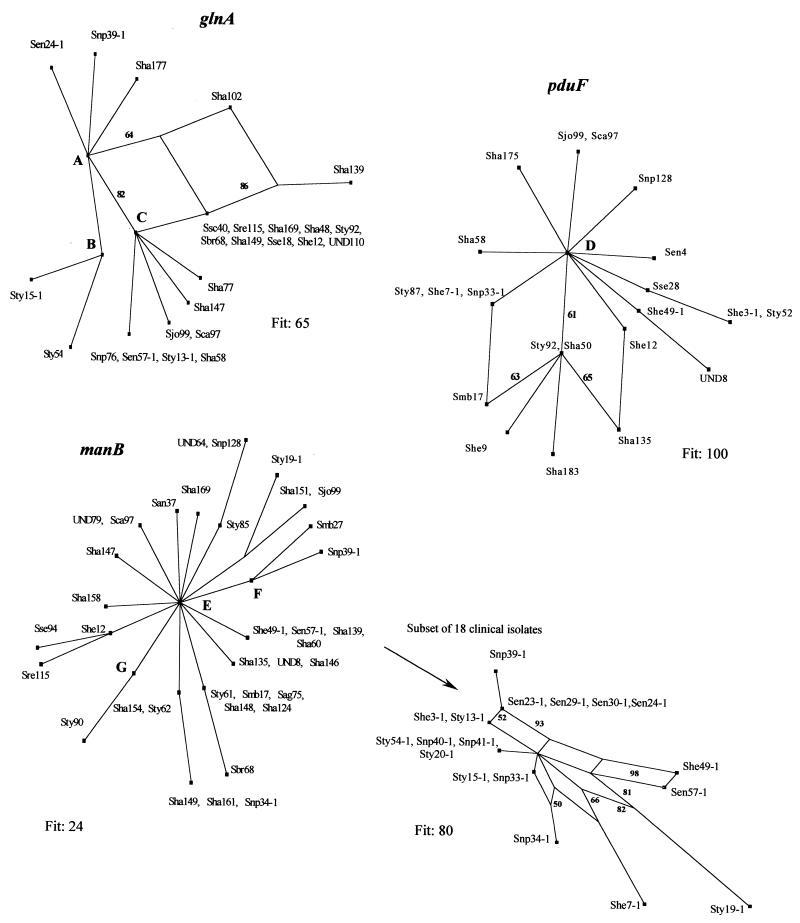
The subset of 107 strains used for the allele and polymorphic site distribution studies was also used to determine (by the split decomposition method) interrelationships among Salmonella strains based on the manB, pduF, and glnA gene fragments (Fig. 4). Splitstree analysis of the glnA and pduF loci revealed two parallelograms for each gene. The fits were 65 and 100, respectively, which indicated that the majority of the information was consistent with the analysis. In contrast, initial Splitstree analysis of the manB locus did not reveal any parallelograms (Fig. 4, bottom left). However, the probability fit was very low (fit, 24), which prompted us to examine two smaller subsets of strains by the Splitstree method. Eighteen clinical strains were included in the first subset, and 15 environmental strains were included in the second subset. Six parallelograms were observed for the first subset, and the fit improved dramatically (the fit increased from 24 to 80) (Fig. 4, bottom right). All of the parallelograms had bootstrapping values greater than 50, and approximately half of them had bootstrapping values greater than 80, which indicated that the data were statistically robust. Similarly, a high fit of 84 (with bootstrapping values as high as 57 and 70) and two parallelograms were observed when the manB loci of 15 environmental isolates (the second subset) were analyzed by the Splitstree method (data not shown).
FIG. 4.
Split graphs based on bootstrap (100 replicates) analyses of the glnA, pduF, and manB gene sequences. The designations at the branches are described in the legend to Fig. 3. A includes the following isolates: San96, Sca93, Sen4, Sen5, Sen23-1, Sen29-1, Sen30-1, Sen34, She49-1, Sha56, Sha60, Sha122, Sha130, Sha135, Sha138, Sha150, Sha151, Sha153, Sha157, Sha158, Sha161, Sha164, Sha165, Sha173, Sha175, Sha176, Sha183, Sha186, Sht20, Smb27, Snp34-1, Snp41-1, Snp128, Sse28, Sse94, Sse95, Sty15, Sty62, Sty86, Sty87, Sty90, Swo44, UND3, UND8, UND64, UND79, UND80, and UND109. B includes the following isolates: Sag75, San37, Sen24, Sha71, Sha124, Sha141, Sha146, Sha148, Sha160, Sha182, She7-1, She9, Ske69, Smb17, Snp33-1, Ste41, Snp40-1, Sty2, Sty6, Sty13, Sty19-1, Sty20-1, Sty31, Sty54-1, and Sty61. C includes the following isolates: Sag129, Sha49, Sha50, Sha131, Sha154, She3-1, Sty52, Sty85, and Sty89. D includes the following isolates: Sag75, Sag129, San37, San96, Sbr68, Sca93, Sen5, Sen23-1, Sen24, Sen24-1, Sen29-1, Sen30-1, Sen34, Sen57-1, Sha48, Sha49, Sha56, Sha60, Sha71, Sha77, Sha102, Sha122, Sha124, Sha130, Sha131, Sha138, Sha139, Sha141, Sha146, Sha147, Sha148, Sha149, Sha150, Sha151, Sha153, Sha154, Sha157, Sha158, Sha160, Sha161,Sha164, Sha165, Sha169, Sha173, Sha176, Sha177, Sha182, Sha186, Sht20, Ske69, Smb27, Snp34-1, Snp39-1, Snp40-1, Snp41-1, Snp76, Sre115, Ssc40, Sse18, Sse94, Sse95, Ste41, Sty2, Sty6, Sty13, Sty13-1, Sty15, Sty15-1, Sty19-1, Sty20-1, Sty31, Sty54, Sty54-1, Sty61, Sty62, Sty85, Sty86, Sty89, Sty90, Swo44, UND3, UND64, UND79, UND80, UND109, and UND110. E includes the following isolates: Sag129, Sen4, Sha48, Sha49, Sha102, Sha131, Sha165, Sha175, Sha182, Sha183, She7-1, Ske69, Snp33-1, Ssc40, Sse28, Sty6, Sty15, Sty15-1, Sty52, Sty89, Sty92, UND109, and UND110. F includes the following isolates: San96, Sca93, Sen23-1, Sen24, Sen24-1, Sen29-1, Sen30-1, Sen34, Sha50, Sha56, Sha77, Sha122, Sha130, Sha138, Sha150, Sha153, Sha157, Sha164, Sha173, Sha176, Sha177, Sha186, She3-1, Sht20, Snp76, Sse95, Ste41, StyI3-1, Sty86, Swo44, UND3, and UND80. G includes the following isolates: Sen5, Sha58, Sha71, Sha141, Sha160, She9, Sty2, Sty13, Sty20-1, Sty31, Sty54, Sty54-1, Sty87, Snp40-1, Snp41-1, and Sse18.
DISCUSSION
PFGE analyses.
Seventy-eight distinct PFGE types were identified among the 232 Salmonella strains whose macrorestriction patterns were analyzed during our study, which supports previous reports (2, 10) concerning the genetic heterogeneity of Salmonella. We found that PFGE was more discriminatory than serotyping for the analysis of Salmonella strains, as PFGE differentiated strains indistinguishable by serotyping. In addition, we found that various serotypes differed in their levels of heterogeneity (as assessed by PFGE typing). Among the five serotypes (serotypes Hadar, Typhimurium, Enteritidis, Heidelberg, and Newport) represented by more than 15 strains in our study, Salmonella serotype Hadar and Salmonella serotype Newport were the most homogeneous and heterogeneous serotypes, respectively (Table 1). Caution is required in the interpretation of these data, however, because the homogeneity of the serotypes assessed by our PFGE typing method may be due to the specificity of our strain collection. For example, Salmonella serotype Hadar was considered the most homogeneous serotype because it included 58 strains clustered in a single PFGE type (type P68) by PFGE typing, which may be the result of the collection of multiple strains of clonal origin during a short period of time rather than an indication of the genetic homogeneity of the serotype.
Our PFGE analyses grouped some of the clinical and environmental strains into distinct clusters (e.g., clusters B and C, respectively; Fig. 2). This is an interesting observation, especially in view of the increasing recognition that some Salmonella serotypes and strains may have an increased virulence potential. In this context, only a few of the more than 2,400 currently recognized Salmonella serotypes (5) are predominantly associated with human salmonellosis, and at least some of these serotypes (e.g., serotypes Typhimurium and Enteritidis) have been reported (33, 34) to have increased pathogenic potentials compared to those of the other serotypes. Moreover, approximately 50% of all Salmonella serotype Enteritidis isolates in the United States have been found to belong to phage type 8, and most of the recent major outbreaks of serotype Enteritidis-associated salmonellosis in the United States have been caused by strains belonging to phage type 4 (20, 30), which has been reported (22) to have increased virulence in mice and, possibly, in humans. Thus, our observation that PFGE clustered some of the clinical (and, presumably, highly pathogenic) and environmental (and, presumably, less pathogenic) strains into distinct genetic clusters may reflect the differences in the pathogenic potentials between the strains in the clinical and environmental clusters. However, the clustering of clinical isolates was limited to strains of a single serotype (nine strains of serotype Newport), which were grouped into six closely related PFGE types. Therefore, it is likely that the observed clustering reflects the clonal origins of the strains rather than their increased pathogenic potentials.
Comparison of PFGE and MLST analyses.
In our study, MLST differentiated strains grouped in a single PFGE cluster and/or type. This finding supports our hypothesis that MLST is more discriminating than PFGE for the typing of Salmonella strains because MLST detects all genetic variations within the amplified gene fragment, whereas PFGE examines only those that are in the cleavage sites for the particular restriction enzyme. PFGE does have the advantage of randomly “probing” the entire genome, whereas MLST only analyzes nucleotides within the targeted gene, a limitation which can be overcome by analyzing multiple genes from various regions of the bacterial chromosome.
In order to compare further the discriminatory abilities of MLST and PFGE, we determined (on trees generated by MLST) the average genetic distances among the strains clustered in distinct PFGE types. In other words, all members or strains of specific PFGE groups were located on the tree generated by MLST, and the average genetic distances among the strains were calculated. If the genetic distances for all members of each of the PFGE groups were zero, PFGE would be considered to describe adequately the genetic variation in these strains. However, the genetic distances were found to be greater than zero, which further confirmed our observation that the discriminatory ability of MLST for the typing of various Salmonella serotypes is greater than that of PFGE. MLST also typed the serotype Kentucky isolates and two Salmonella strains of undetermined serotype that were not typeable by PFGE, which suggests that MLST can be successfully used to type Salmonella strains which cannot be analyzed by the standard PFGE procedure for Salmonella strains.
Clustering of clinical and environmental strains by MLST.
Analyses of the DNA sequences by either the maximum parsimony or the maximum likelihood method revealed manB sequence-based clustering of clinical versus environmental isolates (Fig. 3). In contrast to the cluster of clinical isolates identified by PFGE (Fig. 2, cluster B), the two manB sequence-based clusters of clinical isolates (Fig. 3, clusters A and B) were not limited to a single serotype but instead contained strains of four serotypes (serotypes Newport, Typhimurium, Heidelberg, and Enteritidis) and several PFGE types. Therefore, it is tempting to speculate that the manB gene may be involved in some, as yet unidentified, function related to the pathogenicity of Salmonella and that the substitutions which caused the strains to be grouped into the cluster of clinical isolates are associated with the strains' potentially increased virulence (compared to the virulence of the strains in the environmental clusters). This hypothesis also supports the idea that the division between “virulence” and “housekeeping” genes in Salmonella (and, possibly, other bacteria) may not be as clear-cut as is currently thought. In this context, interruption of the pduF and glnA clusters of Salmonella have previously been reported (8, 25) to confer reduced virulence. However, the role, if any, of the manB gene in the virulence of Salmonella is not clear at this time, and the “critical” substitutions that cause some strains to be grouped into clusters of clinical isolates could not be identified in the region of the manB gene that we sequenced. The nucleotide changes that were responsible for our separation of the clinical isolates from the neighboring environmental isolates are primarily substitutions in the third codon (data not shown), and thus, they are silent. Therefore, it is possible that the critical substitutions are in lineage disequilibrium with the critical mutation(s) that is outside the region that we sequenced (either in the manB gene or in a neighboring gene). The validity of this hypothesis can be addressed by (i) sequencing, for a large number of clinical and environmental strains, additional Salmonella genes immediately downstream and upstream of the manB gene and (ii) performing rigorous in vitro and in vivo virulence studies with various manB mutants.
Recombinational basis of genetic diversity of the loci analyzed by MLST.
The G+C contents of the glnA and manB genes (61 and 63%, respectively; Table 2) were significantly higher than the overall average G+C content of 51% previously reported (41) for S. enterica and E. coli, which suggests that glnA and manB originated in species other than S. enterica and E. coli and that they were introduced into the common ancestor of Escherichia and Salmonella via horizontal gene transfer. On the other hand, the G+C content of the pduF gene was 51%; i.e., it was very similar to the overall average for the species. This observation supports the idea that the pduF gene either was directly inherited by Salmonella and E. coli from the same ancestor or was lost by a common ancestor of E. coli and Salmonella spp. and was reintroduced as a single fragment into the Salmonella lineage from an exogenous source, as suggested previously (26).
The results of the Splitstree analysis suggested that recombinational events played an important role in the evolution of the glnA and pduF genes (Fig. 4). Initial Splitstree analysis did not reveal parallelograms in the manB gene and, thus, evidence of recombination. However, the fit value of 24 was very low, which could be due to the inability of the program to mathematically describe an excessive number of recombinational events within the gene. To test this hypothesis, we repeated the Splitstree analysis with a subset of 18 clinical isolates, and we observed a significant improvement in the fit values (fit, 80). Moreover, six parallelograms were observed (with bootstrapping values as high as 98 and 93) (Fig. 4, bottom right), which suggests that many mutations are involved in the parallel events and that recombinations in the manB gene are frequent. Similar results were observed when a subset of environmental strains was analyzed by Splitstree analysis, which confirms the observation made above and which indicates that recombination in the manB gene is not limited to clinical isolates.
In conclusion, our study is the first peer-reviewed publication concerning MLST of Salmonella, and it demonstrates that MLST has a better discriminatory ability than serotyping and PFGE typing for the typing of various Salmonella strains. The improved discriminatory ability of MLST can be useful for the differentiation of strains involved in food-borne outbreaks of salmonellosis, including strains within the same serotype that are strongly clonal and that may therefore not be differentiable by PFGE typing. Furthermore, our data indicate that, in addition to being a valuable epidemiologic tool, MLST can be useful in suggesting novel putative virulence markers in Salmonella (and, possibly, other species) and may be invaluable for determination of the genetic relatedness among various Salmonella strains and serotypes. Our results also support previous observations (7) that various genes of Salmonella differ in their evolutionary origins and that they evolve at different rates via various evolutionary mechanisms, including recombination. The important implication of this observation is that evolutionary processes within Salmonella—and, most likely, within and/or among other bacterial species—cannot be postulated from data for a single genetic locus (or even a few genetic loci) and that sequencing of multiple genes (or an entire bacterial genome) and the use of various powerful data analysis algorithms are required to gain an improved understanding of the epidemiology and evolution of Salmonella.
Acknowledgments
We thank Keith Jolley for generous assistance with the START program, Ekaterine Chighladze for help with subculturing and cataloguing of the bacterial strains, Durmishkhan Turabelidze for PFGE typing some of the Salmonella strains used in the study, and Siqen Zheng for assistance during the design of some of the MLST primers.
M.K. was supported by an International Training and Research in Emerging Infectious Diseases grant from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. (Erratum, 97:8192, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, and A. Cravioto. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, J. B., C. W. Hedberg, D. J. Boxrud, J. M. Besser, J. H. Wicklund, K. E. Smith, and M. T. Osterholm. 2001. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N. Engl. J. Med. 344:189-195. [DOI] [PubMed] [Google Scholar]
- 4.Boonmar, S., A. Bangtrakulnonth, S. Pornrunangwong, J. Terajima, H. Watanabe, K. I. Kaneko, and M. Ogawa. 1998. Epidemiological analysis of Salmonella enteritidis isolates from humans and broiler chickens in Thailand by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:971-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bopp, C. A., F. W. Brenner, J. G. Wells, and N. A. Strockbine. 1999. Escherichia, Shigella, and Salmonella, p. 459-474. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolkin (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 6.Borrego, J. J., D. Castro, M. Jimenez-Notario, A. Luque, E. Martinez-Manzanares, C. Rodriguez-Avial, and J. J. Picazo. 1992. Comparison of epidemiological markers of Salmonella strains isolated from different sources in Spain. J. Clin. Microbiol. 30:3058-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., K. Nelson, F. S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, K., I. Linetsky, C. Hueser, and A. Eisenstark. 2001. Genetic variability among archival cultures of Salmonella typhimurium. FEMS Microbiol. Lett. 199:215-219. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing, B., and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Ewing, B., and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 15.Fadl, A. A., A. V. Nguyen, and M. I. Khan. 1995. Analysis of Salmonella enteritidis isolates by arbitrarily primed PCR. J. Clin. Microbiol. 33:987-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. J. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Food Safety Inspection Service. 1996. 9 CFR Part 304, et al. Pathogen reduction; hazard analysis and critical control point (HACCP) systems; final rule. Fed. Regist. 61:38806-38989. [Google Scholar]
- 19.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in one day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman-Brenner, F. W., A. D. Stubbs, and J. J. Farmer III. 1991. Phage typing of Salmonella enteritidis in the United States. J. Clin. Microbiol. 29:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillis, D. M., M. W. Allard, and M. M. Miyamoto. 1993. Analysis of DNA sequence data: phylogenetic inference. Methods Enzymol. 224:456-487. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey, T. J., A. Williams, K. McAlpine, M. S. Lever, J. Guard-Petter, and J. M. Cox. 1996. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol. Infect. 117:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huson, D. H. 1998. SplitsTree: analysing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 25.Klose, K. E., and J. J. Mekalanos. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, J., K. Nelson, A. C. McWhorter, T. S. Whittam, and R. K. Selander. 1994. Recombinational basis of serovar diversity in Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rampling, A., J. R. Anderson, R. Upson, E. Peters, L. R. Ward, and B. Rowe. 1989. Salmonella enteritidis phage type 4 infection of broiler chickens: a hazard to public health. Lancet ii:436-438. [DOI] [PubMed]
- 31.Reeves, P. 1993. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 9:17-22. [DOI] [PubMed] [Google Scholar]
- 32.Ridley, A. M., E. J. Threlfall, and B. Rowe. 1998. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarwari, A. R., L. S. Magder, P. Levine, A. M. McNamara, S. Knower, G. L. Armstrong, R. Etzel, J. Hollingsworth, and J. G. Morris, Jr. 2001. Serotype distribution of Salmonella isolates from food animals after slaughter differs from that of isolates found in humans. J. Infect. Dis. 183:1295-1299. [DOI] [PubMed] [Google Scholar]
- 34.Solano, C., B. Sesma, M. Alvarez, T. J. Humphrey, C. J. Thorns, and C. Gamazo. 1998. Discrimination of strains of Salmonella enteritidis with differing levels of virulence by an in vitro glass adherence test. J. Clin. Microbiol. 36:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. G. Morris, Jr., and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stubbs, A. D., F. W. Hickman-Brenner, D. N. Cameron, and J. J. Farmer III. 1994. Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and biotyping. J. Clin. Microbiol. 32:199-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford, D. 2000. PAUP (phylogenetic analysis using parsimony). Sinauer Associates, Sunderland, Mass.
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thong, K. L., Y. F. Ngeow, M. Altwegg, P. Navaratnam, and T. Pang. 1995. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J. Clin. Microbiol. 33:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Threlfall, E. J., and J. A. Frost. 1990. The identification, typing and fingerprinting of Salmonella: laboratory aspects and epidemiological applications. J. Appl. Bacteriol. 68:5-16. [DOI] [PubMed] [Google Scholar]
- 41.Wang, L., and P. R. Reeves. 2000. The Escherichia coli O111 and Salmonella enterica O35 gene clusters: gene clusters encoding the same colitose-containing O antigen are highly conserved. J. Bacteriol. 182:5256-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]