Molecular methods have now become established as accepted methods for the detection of causal agents of infection (viral, bacterial, fungal, and protozoal). In particular, the use of a combination of rRNA genes from bacteria, fungi, and protozoa, i.e., universal or broad-range targets, has become popular for detection. However, the laboratory workup of clinical specimens for universal PCR differs significantly from that for specific PCR in that numerous problems, mainly related to contamination with DNA, are encountered. Therefore, it is the aim of this review to give practical guidance to help laboratory personnel overcome problems associated with the employment of broad-range ribosomal DNA PCR in the detection of bacterial agents, particularly in culture-negative infections.
The traditional basis for the identification of pathogenic and commensal organisms has been their isolation or propagation in the laboratory. Biochemical, morphological, and serological tests usually require growth of the organism. Reliance on these parameters may have significantly limited awareness of true bacterial diversity and is impractical in many situations. The rapidly expanding use of 16S rRNA sequences for phylogenetic, evolutionary, and diagnostic studies offers an opportunity for alternative approaches (3). 16S rRNA genes are found in all bacteria and accumulate mutations at a slow, constant rate; hence, these genes may be used as molecular clocks (18). Highly variable portions of the 16S rRNA sequence contain signatures unique for each bacterium, as well as useful information about the relationships between different bacteria. Alternatively, since 16S rRNA molecules have crucial structural constraints, certain conserved regions of sequence are found in all known bacteria, including the eubacteria. Broad-range PCR primers may then be designed to recognize these conserved bacterial 16S rRNA gene sequences and used to amplify intervening, variable, or diagnostic regions (15, 17). Broad-range ribosomal DNA (rDNA) PCR avoids the need to grow the bacterium and requires no pre-existing phylogenetic information. Extension of this procedure to the study of infected mammalian tissue further avoids the need to purify the bacterium and has led to the identification of previously uncharacterized pathogens in bacterial angiomatosis (12) and in Whipple's disease (3, 4).
In bacteria, there are three genes which make up the rRNA functionality: the 5S, 16S, and 23S rRNA genes. The 16S rRNA gene has historically been most commonly employed; however, more recently, employment of the 16-23S rRNA intergenic spacer region, along with the 23S rRNA gene, has become popular.
Employment of rRNA-based techniques has gained increased popularity as a means of detection of a diverse variety of bacterial targets from several different clinical specimen types. Since the development of PCR within the life sciences in the late 1980s, there has been a gradual shift away from functional to molecular techniques for the identification of organisms, and this has been reflected pro rata in the literature (6, 7, 8, 13, 14). For example, the cumulative total of papers published in this field in 1999 was greater than the combined output for the period from 1990 to 1994. However, broad-range rDNA PCR involves numerous practicalities and complexities, troubles with which have been a constant problem for laboratories involved in the employment of this technique to help detect bacteria from clinical specimens. Therefore, it is the aim of this review to examine the practical problems associated with broad-range rDNA PCR as well as to offer various suggestions to aid in the production of quality data.
CONTAMINATION AND ITS AVOIDANCE
The major practical problem associated with the use of broad-range rDNA PCR is contamination of the assay by exogenous bacterial DNA. The concept of using highly conserved oligonucleotide primers for a variety of genes within the rRNA structure exploits the phylogeny between the eubacteria. However, the use of such primers can be problematical due to the coamplification of contaminating DNA of little or no clinical significance along with that of the bacterial pathogen. This laboratory complication has led to a number of diagnostic laboratories abandoning the idea of adopting broad-range rDNA PCR techniques as part of their diagnostic service. In addition, contamination has given rise to numerous false-positive results which are of no beneficial importance to the patient and indeed may lead to added complications in the clinical interpretation of molecular results. Careful control of contamination from any source is the key to the successful adoption of this technique within the microbiology service.
EVALUATION OF CONTAMINATION HAZARDS THROUGH RISK ASSESSMENT MODELS
Employment of broad-range rDNA PCR is always associated with the risk of amplification of contaminating DNA which is of no clinical significance. As this risk cannot be avoided under normal laboratory conditions, attempts should be made to help quantify the hazards and risks encountered during the complete diagnostic procedure. The most critical step in establishing protocols for broad-range rDNA PCR is performing a risk assessment to examine ways of reducing or eliminating contamination. In this regard, a contamination hazard may be defined as the introduction of contaminating DNA from any source, e.g., from the patient or during end-stage laboratory manipulation. There are two types of hazards, which may be classified as high and low. A high hazard may be defined as large amounts of contaminating DNA entering the diagnostic assay, whereas a low hazard may be defined a small amounts of DNA entering the diagnostic assay. Risk may be defined as the probability of the hazard occurring. These parameters may be further qualified by assigning contamination categories to any given manipulation (Table 1). Given four categories, categories A to D, with decreasing levels of associated risk, all manipulations in category A will mostly result in the introduction of contaminating DNA into the process, whereas those procedures in category D will by and large not result in the introduction of contaminating DNA, although there is still a small probability of this occurring. The goal of performing a risk assessment is to carefully consider all hazards and risks associated with the empirical use of molecular diagnostics under the conditions examined so that the risk can be reduced as much as possible. In addition, the risk assessment should be carried out by a group of workers representing all staff who may potentially interact with (i) the diagnostic process (i.e., ward staff and laboratory personnel) and (ii) the diagnostic environment (i.e., laboratory personnel not directly involved with the processing of the specimen, porters, and maintenance and cleaning staff).
TABLE 1.
Risk assessment categorization of contaminating DNA compromising employment of broad-range rDNA PCR as a diagnostic tool in the detection of bacterial causal agents of infectious disease
| Category | Hazard | Risk | Example of contamination | Corrective action |
|---|---|---|---|---|
| A | High | High | Employment of commercially available blood collection vials containing EDTA | Laboratory production of DNA-free blood collection vials containing EDTA by using molecular-grade water and chemicals |
| B | High | Low | Misuse of pipette, leading to contamination of pipette barrel | Care with pipetting and use of plugged pipette tips |
| C | Low | High | Acquisition of clinical specimen from patient | Education and training of ward staff to avoid collection of commensal skin flora from patient |
| D | Low | Low | Employment of molecular-grade PCR reagents, e.g., Taq polymerase, contaminated with bacterial DNA | Employment of prescreened Taq polymerase to ensure DNA-free status |
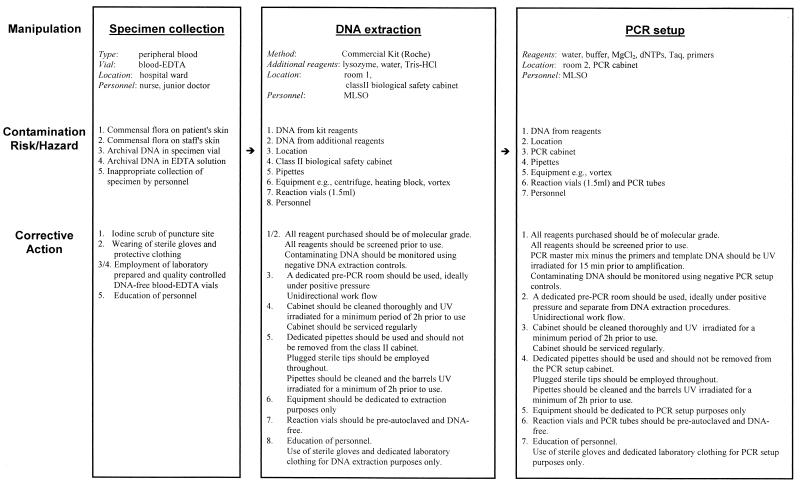
The first step of any such risk assessment should be the defining of the diagnostic procedures through appropriate flow diagrams (an example is shown in Fig. 1). The protocol for the handling and processing of different specimen types requires the generation of a structured flow diagram. Any deviation from the normal procedure requires a reevaluation of the process in terms of additional hazards and/or risks and appropriate control measures established to compensate for these added hazards and/or risks. As each laboratory has its own unique working practices and environment, no single strategy for hazard analysis and critical control point correction is appropriate for adoption by all working laboratories, and all laboratories should therefore carry out their own risk assessments and adopt appropriate control strategies commensurate with their calculated level of hazard. Only when all hazards and risks are identified can appropriate control measures be adopted either (i) to reduce the hazard and/or risk or (ii) to eliminate the hazard and/or risk. The available mechanisms for achieving either the former or the latter are discussed in detail below.
FIG. 1.
Example of a flow chart detailing the manipulations, the contamination hazards and risks, and the corrective action involved for broad-range 16S rDNA PCR detection (in peripheral blood) of bacterial causal agents of infectious diseases. Blood-EDTA vials are blood collection vials containing EDTA. dNTPs, deoxyribonucleoside triphosphates; MLSO, medical laboratory scientific officer.
Clinical specimen. (i) Specimen type.
Broad-range rDNA PCR amplification may be performed only on clinical specimens which are normally regarded as sterile, e.g., blood, cerebrospinal fluid (CSF), pleural fluid, or sterile biopsy material. Employment of this technique with specimens from nonsterile sites, e.g., feces, skin, sputum, or nonsterile biopsy material, should be avoided because the diverse variety of the commensal flora at these sites would complicate the PCR analysis. Amplification of DNA from such sites would yield numerous PCR amplicons for each of the taxa present in the clinical specimen, thus necessitating the separation of such amplicons by further procedures, such as cloning, prior to sequence analysis. For a specimen from a nonsterile site, it would be appropriate to use a more directed molecular approach employing PCR assays specific for a particular gene target, one that is unique to a given organism, e.g., the recA locus for the detection of Burkholderia cepacia in patients with cystic fibrosis (10).
(ii) Collection of specimen.
Like conventional assays, molecular assays require that aseptic precautions be taken in the collection of clinical specimens. However, due to the increased sensitivity of molecular assays compared to that of culture assays, additional stringency should be employed in the collection of specimens for any downstream molecular workup, e.g., in the collection of venous blood, the puncture site should be prewashed with iodine. All personnel involved in the collection of clinical specimens, including medical and nursing staff, as well as phlebotomists, should be educated with respect to the added precautions needed to avoid contamination.
Care should be taken with all equipment and instruments which can potentially come into contact with the clinical specimen being examined by PCR, as these instruments, though considered sterile, can contain DNA from nonviable cells which have been killed in the sterilization process. Recently Keay et al. (5) demonstrated that sterile cold-cup biopsy forceps were a source of Escherichia, Propionobacterium, Stenotrophomonas, and Pseudomonas DNA contamination and that it was therefore inappropriate to use PCR to look for the presence of bacterial pathogens in tissue specimens that had been procured with these instruments.
Sufficient clinical material should be taken to allow for both culture and molecular analysis, and, where possible, two specimens should be taken simultaneously. In cases where limited clinical material can be collected (e.g., specimens of CSF or heart valve tissue), analysis should initially be molecular to avoid potential contamination that may arise from the use of nonsterile plasticware during conventional analysis, e.g., the use of nonsterile pipette tips in the determination of glucose and protein concentrations and white blood cell counts in CSF specimens from meningitis patients.
Specimen collection vials are a source of contaminating DNA. Such DNA may be attributed to both the plasticware and the transport physiological milieu. Previous studies have demonstrated the presence of DNA from nonviable environmental pseudomonads in standard blood collection vials containing EDTA that are used on hospital wards. It is therefore necessary to eliminate this potential source of contamination through the adoption of preparing sterile DNA-free EDTA-treated blood collection vials for the purpose of clinical samples which require broad-range molecular analysis. To help minimize such contamination, the use of high quality virgin tissue-culture plasticware, along with EDTA and water which are DNA free, is recommended. All prepared batches should be screened for the presence of contaminating DNA before distribution to the wards.
Another source of contaminating bacterial DNA is commercially available blood culture material for use with continuously monitoring automated blood culture instruments, e.g., the BacTec and BacT/Alert systems. Such blood culture material is considered sterile; i.e., it does not contain any culturable organisms. Various studies have demonstrated the presence of contaminating DNA in blood culture material from different suppliers. The origin of this bacterial DNA is dependent on both the supplier and the batch (lot) number. Several sources of bacterial DNA have been identified by molecular sequence analysis and include DNA from Lactococcus lactis and Bacillus coagulans (9), as well as a Streptococcus sp. (2).
Organization of the working environment.
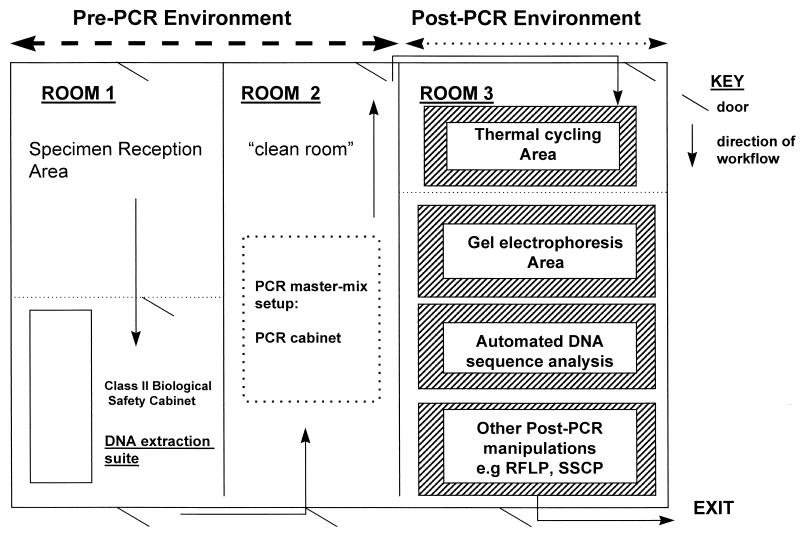
One of the fundamental requirements of broad-range rDNA PCR is segregation of the laboratory work areas to avoid the amplification of artifactual contaminating DNA. At minimum, the areas where pre- and post-PCR manipulations are performed should be physically separated. Pre-PCR manipulations include all procedures prior to thermal cycling, and, consequently, post-PCR manipulations include all stages downstream and including thermal cycling (Fig. 2). However, the ideal physical arrangement would allow for two separate pre-PCR rooms, with one room being dedicated to clinical specimen reception and genomic DNA extraction and the second room being classed as the clean room, where the PCR master mixes are set up (Fig. 2). Within each room, manipulations with respect to broad-range and specific PCR analyses should be physically segregated. Such segregation may be implemented at two levels: (i) infrastructural segregation and (ii) local containment. Infrastructural segregation involves physical separation by means of a partitioning wall or a solid wall. Additionally, segregated facilities should be geographically sited in an area with minimum human traffic.
FIG. 2.
Proposed layout of laboratory space associated with broad-range 16S rDNA PCR for detection of bacterial causal agents of infectious diseases. RFLP, restriction fragment length polymorphism; SSCP, single-stranded conformational polymorphism.
Local containment involves the employment of biological safety cabinets for initial specimen disinfection and DNA extraction. Such cabinets should not be used for PCR setup, as this procedure should be conducted apart from the aforementioned disinfection and extraction to minimize contamination. A recent innovation with laboratory equipment manufacturers has been the PCR setup cabinet, or so-called dead-air box. These cabinets have been designed exclusively for PCR setup purposes and should not be jointly employed for DNA extraction manipulations. Such cabinets vary in the complexity of their specifications from simple dead-air boxes to those with HEPA-filtered circulating air (equivalent to airflow within a class II biological safety cabinet, e.g., the Omni PCR cabinet (Microflow Ltd. Weston-super-mare, United Kingdom), as well as UV light facilities for internal decontamination of the work area, air, pipette barrels, and PCR master mixes. Both the extraction and PCR cabinets should intrinsically minimize the opportunity for specimen contamination. For specimen protection, class II safety cabinets, in which filtered air is recirculated within the cabinet through HEPA filters, should be employed, as opposed to class I safety cabinets, in which ambient air is drawn in to the cabinet to maximize staff safety. However, the use of the former type of cabinet may therefore create certain health and safety concerns, e.g., during extraction of DNA from blood or biopsy material containing Mycobacterium tuberculosis. It is recommended that the class II and PCR setup cabinets each be equipped with a UV lamp in order to degrade any exogenous DNA prior to PCR amplification. Recently, we have noted the importance of maintaining the working efficiency of such cabinets, as these may become a source of contamination if the HEPA filters are not serviced regularly. Furthermore, the efficiency of UV light production should be monitored regularly.
Risk from laboratory personnel.
The hazards associated with laboratory personnel may originate from the contamination of specimens by staff dedicated to molecular diagnostic manipulations, whereby common skin contaminants, including coagulase-negative staphylococci, may be introduced. In order to reduce the risk of this type of contamination occurring, staff should wear sterile rubber or latex gloves as well as separate white coats, respectively dedicated to broad-range DNA extraction, master mix setup, and post-PCR manipulations.
Consumable reagents and plasticware.
Contamination can enter the broad-range diagnostic protocol with the addition of each reagent (6) during the DNA extraction and amplification procedures. Specifically, we have noted four problem areas for bacterial DNA contamination, as follows: (i) lytic enzymes used for extraction of DNA from yeasts and bacteria, (ii) oligonucleotide primers, (iii) Taq polymerase (11), and (iv) water. Most manufacturers of consumable reagents do not guarantee their products to be DNA-free, although most would give an assurance that they are sterile (i.e., contain no viable organisms) and even DNase-free (for most molecular reagents). Consequently, these reagents may constitute a risk of contamination that can be eliminated through prior screening of individual reagents before employment in diagnostic assays. Plasticware used during prescreening should be of high quality and should not be reused in molecular assays. For pipetting purposes, it is important that three dedicated sets of pipettes exist for DNA extraction, master mix setup, and post-PCR manipulations, respectively, where the former two sets of pipettes should be UV irradiated for at least 2 h after use. In addition, it is imperative that only pipette tips which are filtered are employed during these manipulations.
MANAGEMENT OF CONTROLS
Successful employment of broad-range rDNA PCR in the detection of causal agents of infectious disease is critically dependent on both the quantity and the quality of controls associated with the assay. It should be noted that, for each PCR-based test, sensitivity should be evaluated for each specimen type, prior to routine implementation. It is vital that several negative and positive controls be set up during each diagnostic run. Negative and positive controls should include (i) a DNA extraction control, (ii) a PCR setup control, and (iii) a PCR amplification control.
For DNA extraction purposes, the positive control should include a clinical specimen(s) artificially spiked with an organism, e.g., blood culture spiked with Escherichia coli. In cases where a true-positive clinical tissue specimen may be difficult to mimic, internal positive controls, such as amplification of the β-globin gene following DNA extraction, as previously described (8), may be employed. With respect to PCR controls, the positive control should be bacterial DNA extracted from a pure culture. Ideally, the positive control should include two components, namely, (i) a specimen generating a weak signal, due to low copy numbers of target, and (ii) a specimen generating a strong signal, due to high copy numbers of target. Employing these controls, especially the negative controls, makes it easier to identify the point of contamination within the diagnostic assay, e.g., to verify that the DNA extraction procedure was contamination free but that contamination might have occurred during PCR setup.
Positive controls, particularly those included in the DNA extraction procedures, are also important because they serve to identify possible inhibition of the PCR due to inhibitory agents in the biological specimen which coelute with extracted DNA, e.g., sodium polyanetholesulfonate in blood culture material (2, 8). For a comprehensive review on PCR inhibition with respect to biological specimens, see the study by Wilson in reference 16.
More recently, technological advances in the equipment used for PCR have occurred, and the newer resulting processes have been more commonly applied in the diagnosis of infectious diseases. Although such advances, including real-time PCR, may have increased sensitivity compared with conventional PCR, there are several problems with using these platforms in conjunction with broad-range 16S rDNA PCR. Recently Corless et al. (1) described numerous problems associated with the use of the Taqman system and 16S rDNA PCR. These workers concluded that the added sensitivity meant that contaminants associated within the entire diagnostic assay, were more easily detected, though they were of no clinical significance, and that one potential way to overcome such contamination issues was to use ultraclean reagents and plasticware.
In conclusion, the successful implementation of broad-range rDNA PCR targeting highly conserved loci within the 16S rRNA gene presents a number of complexities and challenges. Conceptually and physically, broad-range rDNA PCR should be considered as a separate protocol in molecular diagnostics rather than as an add-on to specific PCR. Success can be achieved through careful management of the working environment and reagents, which may be accomplished more easily by identifying the risks and hazards of contamination through a structured risk assessment approach.
Acknowledgments
B.C.M, X.J., and J.E.M. are supported by the Primary Immunodeficiency Association, Action Cancer, the Cancer Research Campaign, and the Department of Health and Social Services (Belfast, Northern Ireland).
REFERENCES
- 1.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fredricks, D. N., and D. A. Relman. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J. Clin. Microbiol. 36:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredricks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-486. [DOI] [PubMed] [Google Scholar]
- 4.Gao, S. J., and P. S. Moore. 1996. Molecular approaches to the identification of unculturable infectious agents. Emerg. Infect. Dis. 2:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keay, S., C. O. Zhang, B. R. Baldwin, R. B. Alexander, and J. W. Warren. 1998. Polymerase chain reaction amplification of bacterial 16S rRNA genes from cold-cup biopsy forceps. J. Urol. 160:2229-2231. [DOI] [PubMed] [Google Scholar]
- 6.Kotilainen, P., J. Jalava, O. Meurman, O. P. Lehtonen, E. Rintala, O. P. Seppälä, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier, A., D. H. Persing, M. Finken, and E. C. Bottger. 1993. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J. Clin. Microbiol. 31:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millar, B., J. Moore, P. Mallon, J. Xu, M. Crowe, R. McClurg, D. Raoult, J. Earle, R. Hone, and P. Murphy. 2001. Molecular diagnosis of infective endocarditis—a new Duke's criterion. Scand. J. Infect. Dis. 33:673-680. [DOI] [PubMed] [Google Scholar]
- 9.Millar, B. C., X. Jiru, J. E. Moore, and J. A. P. Earle. 2000. A simple and sensitive method to extract bacterial, yeast and fungal DNA from blood culture material. J. Microbiol. Methods 42:139-147. [DOI] [PubMed] [Google Scholar]
- 10.Moore, J. E., B. C. Millar, X. Jiru, J. McCappin, M. Crowe, and J. S. Elborn. 2001. Rapid characterization of the genomovars of the Burkholderia cepacia complex by PCR-single-stranded conformational polymorphism (PCR-SSCP) analysis. J. Hosp. Infect. 48:129-134. [DOI] [PubMed] [Google Scholar]
- 11.Petershofen, E. K., R. Fislage, R. Faber, H. Schmidt, W. K. Roth, and E. Seifried. 2000. Detection of nucleic acid sequences from bacterial species with molecular genetic methods. Transfus. Sci. 23:21-27. [DOI] [PubMed] [Google Scholar]
- 12.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 13.Rolph, H. J., A. Lennon, M. P. Riggio, W. P. Saunders, D. MacKenzie, L. Coldero, and J. Bagg. 2001. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 39:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotha, R., T. Hanck, W. Konig, and B. Konig. 2001. Rapid ribosequencing—an effective diagnostic tool for detecting microbial infection. Infect. 29:12-16. [DOI] [PubMed] [Google Scholar]
- 15.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]