Abstract
In a comparison of the Directigen and VIDAS respiratory syncytial virus antigen detection assays with viral culture, the sensitivity, specificity, positive and negative predictive values, and testing efficiency were 86, 93.1, 82.7, 94.6, and 91.2% for Directigen; 96.1, 90.8, 80.3, 98.3, and 92.3% for VIDAS; and 88.2, 100, 100, 95.7, and 96.8% for viral culture, respectively.
Respiratory syncytial virus (RSV) is an important respiratory viral pathogen seen primarily in very young children, usually under the age of 1 year (5). Peak RSV season in the United States generally extends from October through the end of March, with sporadic cases detected year-round (2). Infection with this virus may predispose children to develop reactive airway disease and may also cause severe disease in the setting of preexisting childhood asthma (10). In the hospital setting, RSV may be efficiently transmitted from patient to patient if appropriate infection control measures are not undertaken (6). Early diagnosis of RSV is very important if the cohorting of patients is to be effective (4), and it helps to dictate appropriate infection control precautions to be employed when infected children are admitted (3, 7). This early diagnostic information may also be used by clinicians to direct antibiotic use in patient management (1). Although RSV is culturable in the clinical laboratory, culture techniques may take days or weeks to detect the virus (1, 9). Children have oftentimes been discharged before culture results were finalized. Additionally, inadequate specimen handling often leads to problems in recovering the virus from specimens due to its labile nature. Success in isolating the virus also depends upon the cell line used and the type of culture format employed (shell vial versus tube culture) (8, 9). Antigen detection techniques have therefore become more widely employed for the rapid detection of RSV disease. This study compared the performance of two RSV antigen detection assays, Directigen RSV ColorPac and VIDAS automated enzyme immunoassay (EIA), with culture for 186 sequential clinical respiratory specimens tested at the University of Kentucky Hospital. Specimens included nasopharyngeal aspirates, nasal washes, suctioned sputa, endotracheal aspirates, and throat swabs from both children and adults. Raw specimens were transported to the laboratory on ice and either processed immediately or refrigerated until testing and culture. The Institutional Review Board approved the protocol for compliance with human subject safety requirements.
Viral culture was performed by inoculating clinical specimens onto monolayers of Hep-2 and rhesus monkey kidney cells (BioWhittaker, Walkersville, Md., or Viromed Laboratories, Inc., Minneapolis, Minn.) in the standard tube culture format. Cultures were incubated at 33 ± 2°C for at least 10 days, with review every day for the first week and three times per week thereafter. RSV was detected by blind antibody fluorescence staining at day 4 of incubation in the rhesus monkey kidney cells or after the detection of the characteristic cytopathic effect in Hep-2 culture, which was confirmed by antibody fluorescence staining (Intracel, Issaquah, Wash.). For the purposes of this study, blind fluorescence antibody staining was also performed on all specimens with positive EIA results prior to finalizing cultures if the culture was not positive by day 10. Terminal fluorescence antibody staining was not performed on the cultures if both EIAs were negative.
Antigen detection was performed by a manual Directigen RSV assay (BD Microbiology Systems, Cockeysville, Md.) according to the manufacturer's guidelines and on a Mini VIDAS by the VIDAS automated enzyme-linked fluorescent immunoassay (BioMerieux Vitek Inc., Hazelwood, Mo.). The Directigen assay is an EIA that utilizes visual detection of peroxidase staining in the ColorPac solid matrix. Total hands-on and reporting time for the Directigen assay was about 15 min. The Mini VIDAS instrument provides automated enzyme-linked fluorescence detection for RSV antigen in clinical specimens. Specimens are inoculated into a solid-phase receptacle containing reagents in separate wells. All assay steps are performed automatically by the instrument, with the final well acting as a cuvette through which antigen is detected by fluorescence staining. Although the total hands-on time for the assay was similar to that for the Directigen assay (about 15 min), the total testing time with instrument incubations resulted in a turnaround time for final reporting of results of about 3 h. Because of staffing and workflow issues, testing by both methodologies was batched one to two times each day.
The interpretation of results followed the manufacturer's guidelines. The presence of a purple triangle in the test area together with a purple internal control dot was interpreted as positive for the Directigen ColorPac assay. Negative samples demonstrated a purple control spot without any staining within the testing triangle. Specimens were reported as equivocal if the internal control failed to stain or if the testing matrix surrounding the testing triangle demonstrated any nonspecific staining. For the VIDAS assay, relative fluorescence values (RFVs) were used to establish cutoff absorbencies for positive and negative results. Background fluorescence was subtracted from each specimen reading. The sample test strip RFV was divided by the sample reference strip RFV. If the resultant ratio was less than 1.40, the specimen was interpreted as negative. If the ratio was greater than or equal to 1.80, the specimen was considered positive. Ratios between these two values were considered equivocal.
For statistical analysis, all positive cultures were considered true positives. In addition, specimens with positive results by both antigen detection assays were also considered positive, regardless of the culture result. The presence of a single positive antigen test result was considered a false positive for that assay if the specimen was negative by the other assays.
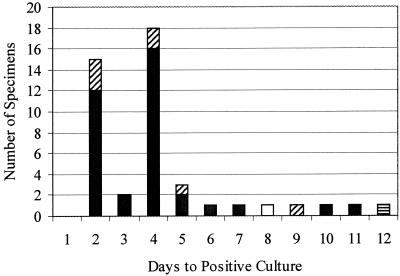
The results of the diagnostic testing are summarized in Table 1. Thirty-six specimens were positive for RSV by culture and both antigen detection assays. An additional seven specimens were positive by culture and the VIDAS assay, one specimen was positive by culture and Directigen, and a single specimen was positive by culture only. The length of time to viral growth in culture ranged from 2 to 12 days (Fig. 1). Fifteen specimens were detected on the second day of culture by the cytopathic effect alone. On day 4 of culture, an additional 18 specimens (40.9% of all positives) were positive by blind fluorescence antibody staining. By day 7 of culture, 90.9% of all positive culture results were detected. Since culture and antigen detection assays are dependent, at least in part, upon the viral load in the direct specimen, it was hypothesized that discrepant EIA results would be seen for the specimens with delayed positive results in culture. However, six of the seven cultures that were positive by VIDAS but not by Directigen were specimens producing positive cultures within the first week after inoculation. The one specimen that was positive by Directigen but negative by VIDAS was positive at day 12 of culture, while the specimen that was positive by culture only was detected at day 8 of culture, weakly supporting the hypothesis. Six more specimens were considered true positives, having been detected by both of the antigen detection assays despite testing negative by culture. Nine specimens were positive by Directigen only, while 12 were positive by VIDAS only. Of the six specimens with equivocal results by VIDAS, four were negative by Directigen and culture while two were positive by Directigen but negative for RSV by culture. One specimen that was equivocal by VIDAS and one false positive were presumptively positive for rhinovirus (demonstrating the characteristic cytopathic effect on fibroblast cells), while two of the false positives grew adenovirus and one grew parainfluenza type 3. Of the five specimens with equivocal results by Directigen, four were negative by VIDAS and culture while one was positive by both. One of these Directigen equivocals was presumptively positive for rhinovirus, and one grew herpes simplex virus type 1. Additionally, three of the false positives by Directigen were presumptively positive for rhinovirus in culture. The overall rate of RSV detection in the clinical specimens analyzed was 51 out of 186, or 27.4%. Limited demographic information was available for 63 patients with RSV-positive cultures and/or positive EIA test results. The majority of patients (84.1%) were under the age of 2 years, 12.7% were 2 to 13 years old, and two patients positive for RSV were adults. The specimen sources were known for these same 63 patients. Suctioned sputa or endotracheal aspirates made up the majority of specimens (77.8%), while 7.9% were nasopharyngeal aspirates, 12.7% were nasal washings, and 1.5% were bronchoalveolar lavage specimens making up the remaining positive specimens. A total of 82% of specimens positive by culture and VIDAS and 77% of those positive by Directigen came from the suctioned sputa and/or endotracheal specimens. The pattern of false-positive and false-negative results correlated neither with specimen type nor patient age.
TABLE 1.
Performance characteristics of RSV culture and antigen detection assays
| Method | No. of indicated type of result
|
||||
|---|---|---|---|---|---|
| True positive | False positive | True negative | False negative | Equivocal | |
| Culture | 45 | 0 | 135 | 6 | 0 |
| VIDAS | 49 | 12 | 119 | 2 | 4 |
| Directigen | 43 | 9 | 122 | 7 | 5 |
FIG. 1.
Number of days until culture detection of RSV in clinical specimens. The majority of specimens that were positive by culture were also positive by both Directigen and VIDAS (filled bars). Seven cultures were positive only by VIDAS (diagonal stripes), while one specimen was positive only by Directigen (horizontal stripes). A single culture was negative by both EIAs (open bar).
The diagnostic sensitivity, specificity, positive and negative predictive values, and overall testing efficiency were calculated for each assay by applying Bayes' theorem (11) (Table 2). The VIDAS RSV antigen detection assay provided the best sensitivity (96.1%) and negative predictive value (98.3%) of the two antigen detection tests compared. The VIDAS assay produced 12 false positives, however, resulting in a specificity of only 90.8%. Rhinovirus, adenovirus, and/or parainfluenza type 3 were cultured out of several of the false-positive and equivocal specimens from the VIDAS assay. Although culture had, by definition, 100% specificity and positive predictive value and an excellent overall testing efficiency (96.8%) and negative predictive value (95.7%), it missed six specimens that were positive by both of the antigen detection assays. These specimens were presumed to be true positives despite the negative culture due to the well-known labile nature of the virus. A delay in specimen transport or mistakes in storage of the specimen prior to its arrival in the lab could produce false-negative cultures even if the patient was actually infected. In addition, the turnaround time for culture (range, 2 to 12 days) makes this an inefficient testing modality. STAT testing for the detection of RSV is often required to help physicians determine appropriate patient treatment, cohorting, and infection control practices (1, 7). In this study, the earliest the virus was detected in culture was at 2 days, giving culture the worst turnaround time after the antigen detection assays, which were completed within 24 h of specimen receipt. Another study looking at the performance of culture compared to rapid antigen detection detected a similar 2-day minimum for culture, even when shell vial culture was used. These authors concluded that culture provided little useful information to assist in patient management due to the prolonged time required to obtain results (1).
TABLE 2.
Further performance characteristics of RSV culture and antigen detection assays
| Method | Test sensitivity (%) | Test specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Testing efficiency (%)a |
|---|---|---|---|---|---|
| Culture | 88.2 | 100 | 100 | 95.7 | 96.8 |
| VIDAS | 96.1 | 90.8 | 80.3 | 98.3 | 92.3 |
| Directigen | 86 | 93.1 | 82.7 | 94.6 | 91.2 |
Testing efficiency is defined as the total number of correctly assigned testing values (true positives and true negatives for each assay) divided by the number of specimens tested (including those with equivocal test results).
The Directigen assay had the worst performance of the three assays in terms of sensitivity, positive and negative predictive values, and overall testing efficiency. The overall specificity (93.1%) was better than that of the VIDAS assay, however, and the turnaround time of this test was much better than that of culture with STAT testing capabilities. Several of the Directigen false positives or equivocals were presumptively positive for rhinovirus, suggesting possible cross-reactivity of this assay with these viruses. This conclusion is made less likely by the fact that the manufacturer had evaluated the kit for possible cross-reactivity with five rhinoviral strains and detected no cross-reactivity. The possibility remains that these specimens represent coinfection of patients with both RSV and rhinovirus.
In conclusion, the VIDAS RSV antigen detection assay was the more sensitive of the two antigen detection methods tested, but it had a significant number of apparent false positives. Although the Directigen assay was the more specific of the antigen detection assays tested in this study, the sensitivity of the assay was only 86%. Either assay could have led to inappropriate cohorting or patient management in a significant number of cases (14 for VIDAS and 16 for Directigen). Although culture ultimately identified 180 of the 186 specimens tested, these results would have been available so late in the patient's disease course that the information would have had no impact on patient management for most, if not all, of the patients.
Acknowledgments
Thanks to Marilyn Kenley, Jan Davis, Louise Morgan, and Beth Chaney for their expert management of the virology specimens tested in this study.
REFERENCES
- 1.Adcock, P. M., G. G. Stout, M. A. Hauck, and G. S. Marshall. 1997. Effect of rapid viral diagnosis on the management of children hospitalized with lower respiratory tract infection. Pediatr. Infect. Dis. J. 16:842-846. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2000. Respiratory syncytial virus activity, 1999-2000 season. Morb. Mortal. Wkly. Rep. 49:1091-1093. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1997. Guidelines for prevention of nosocomial pneumonia. Morb. Mortal. Wkly. Rep. 46(RR-1):1-79. [Google Scholar]
- 4.Dogherty, J. A., D. S. Brookfield, J. Gray, and R. A. McEwan. 1998. Cohorting of infants with respiratory syncytial virus. J. Hosp. Infect. 38:203-206. [DOI] [PubMed] [Google Scholar]
- 5.Glezen, W. P., and F. W. Denny. 1973. Epidemiology of acute lower respiratory diseases in children. N. Engl. J. Med. 288:498-505. [DOI] [PubMed] [Google Scholar]
- 6.Hall, C. B. 1983. The nosocomial spread of respiratory syncytial viral infections. Annu. Rev. Med. 34:311-319. [DOI] [PubMed] [Google Scholar]
- 7.Karanfil, L. V., M. Colon, K. Lykens, C. F. Masters, M. Forman, M. E. Griffith, T. R. Townsend, and T. M. Perl. 1999. Reducing the rate of nosocomially transmitted respiratory syncytial virus. Am. J. Infect. Control 27:91-96. [DOI] [PubMed] [Google Scholar]
- 8.Meqdam, M. M., and G. K. Nasrallah. 2000. Enhanced detection of respiratory syncytial virus by shell vial in children hospitalized with respiratory illnesses in northern Jordan. J. Med. Virol. 62:518-523. [DOI] [PubMed] [Google Scholar]
- 9.Microbiology Resource Committee of the College of American Pathologists. 2000. Surveys 2000: specimen VR1-14, vol. VR1-C, p. 2. College of American Pathologists, Chicago, Ill.
- 10.Tuffaha, A., J. E. Gern, and R. F. Lemanske. 2000. The pathobiology of asthma: implications for treatment. Clin. Chest Med. 21:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkel, P., and B. E. Statland. 1991. Interpreting laboratory results: reference values and decision making, p. 49-76. In J. B. Henry (ed.), Clinical diagnosis and management by laboratory methods, 18th ed. W. B. Saunders Co., Philadelphia, Pa.