Abstract
We herein report one case of culture-negative infectious endocarditis (IE) where the organism, Granulicatella elegans, was identified by molecular analysis using broad-range PCR primers complementary to the 16S rRNA gene on the removed valve. The results and utility of this method are discussed.
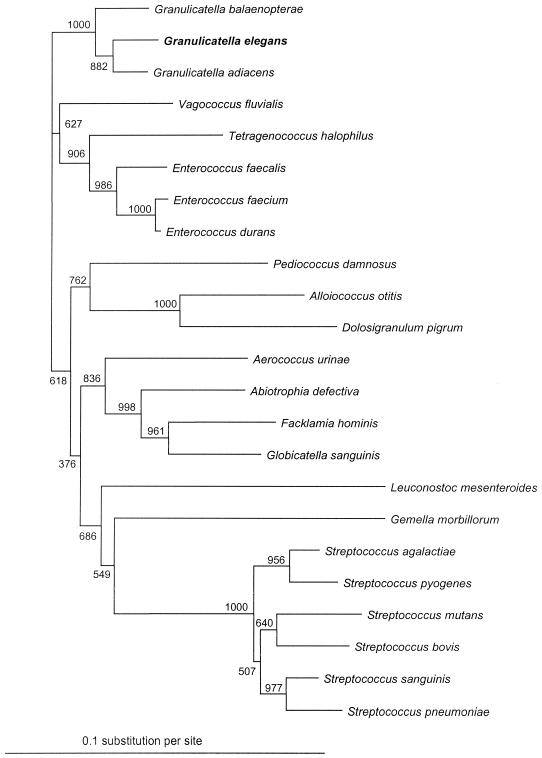
Infectious endocarditis (IE) remains a life-threatening disease. The bacteremia associated with IE is usually low grade and continuous; therefore, blood culture, which usually allows the recovery of the etiologic agent, is considered the most important of the available laboratory diagnostic tests (2). The negative culture results in the classification of such cases into culture-negative endocarditis, which comprises various conditions. Uremia, right-sided or mural endocarditis, prosthetic valve endocarditis, or antibiotic therapy prior to blood culture or valvular surgery may account for negative results (2, 5), but fastidious microorganisms may also be the etiologic agents of culture-negative endocarditis. Among fastidious microorganisms, nutritionally variant streptococci (NVS) were originally described by Frenkel and Hirsch in 1961 (6) as a new type of streptococci exhibiting satellitism around colonies of other bacteria. In 1995, a new genus, Abiotrophia, was created (9). Since then, Collins and Lawson (4) have proposed that three species members of the genus Abiotrophia be reclassified in a new genus, Granulicatella. Granulicatella elegans is the most fastidious among this group of bacteria requiring l-cysteine hydrochloride. By amplification and sequencing of the 16S rRNA through use of broad-spectrum primers (5, 7, 11) (Fig. 1), we were able to identify G. elegans from the valve of a patient with blood culture-negative IE.
FIG. 1.
Dendrogram representing phylogenetic relationships among members of the Streptococcaceae. The tree was derived from a 1,304-bp fragment of the 16S rRNA gene and was constructed by using the neighbor-joining method. Bootstrap values, expressed as a percentage of 1,000 replications, are given at the branching point.
A 29-year-old man was admitted to the Timone hospital in Marseille, France, presenting with clinical signs of acute endocarditis. He complained of fever, anorexia, and weight loss. Different historical data were recorded, including a bicuspid aortic valve, a recent dental procedure 3 months beforehand, and an episode of fever and pneumonia 1 month beforehand, which was treated by amoxicillin and clavulanic acid for 15 days. On admission, the leukocyte count was 7, 650/mm3, the hemoglobin level was 10.8/100 ml, and the erythrocyte sedimentation rate was 37 mm/h. The echocardiography (multiplane transesophageal echocardiography) showed two vegetations on aortic native valves measuring 14 mm at the maximum. Three blood cultures obtained before therapy were negative. The patient was empirically treated with intravenous amoxicillin (2 g every 4 h) and with intravenous gentamicin (3 mg/kg of body weight once a day). Cardiac surgery was performed after 5 days because of heart failure. The aortic valve was replaced with a bioprosthetic valve, and the antimicrobial therapy was continued for a further 30 days. The patient was released from the hospital without fever.
The cultures of the removed valve on Columbia sheep blood agar (reference 43041; BioMerieux, Marcy-l'Etoile, France) and on Chocolate PolyVitex agar (reference 43101; BioMerieux) were negative. Histologic examination of the removed valve was performed, and findings were compatible with the diagnosis of IE. To detect possible bacteria in valvular tissue, PCR incorporating broad-range primers for amplification of the16S rRNA gene was performed. DNA was extracted using Qiagen columns (QIAamp tissue kit; QIAGEN, Hilden, Germany) according to the manufacturer's instructions. PCR amplification using broad-range 16SrRNA gene primers 536f (5′-CAG CAG CCG CGG TAA TAC-3′) and 1050r (5′-CAC GAG CTG ACG ACA-3′) and sequencing and purification of PCR products were performed as previously described (10). After amplification and sequencing of the 16SrRNA gene of this isolate, 544 bp was assembled and the sequence was compared with DNA sequence databases using the program BLAST 2.0 (National Center for Biotechnology Information) and showed 100% homology with the 16S rRNA sequence of G. elegans (GenBank accession no. 3123333) (Fig. 1). The other more closely related species were Granulicatella balaenopterae and Granulicatella adiacens with 98% homology (GenBank accession nos. 4775262 and 4127262, respectively). As this bacterium has never been sequenced in our laboratory before, a contamination is unlikely.
G. elegans is an NVS originally isolated and described by Roggenkamp et al. (12, 13). Cultivation and identification of the different species of NVS are often difficult and can result in misidentification. Taxonomic studies based on DNA-DNA hybridization and sequence analysis of 16S rRNA demonstrated that the genus Abiotrophia (1, 9, 12) was not monophyletic and that Abiotrophia elegans should be reclassified in a new genus, Granulicatella, as G. adiacens, Granulicatella para-adiacens, and G. balaenopterae (3, 4). The genus Abiotrophia should be restricted to Abiotrophia defectiva (4). G. elegans is a possible pathogen in patients with culture-negative endocarditis and has been reported as such. Ten cases of IE caused by G. elegans have been reported in the literature. Among patients with Granulicatella IE reported by Christensen et al. (3), 25 were identified as G. adiacens and 1 as G. elegans. Kanamoto et al. have recently (8) reported 45 cases, including 9 strains of A. defectiva, 15 of G. adiacens, 13 of G. para-adiacens, and 8 of G. elegans. Another case of endocarditis by G. elegans was reported by Roggenkamp et al. (13). All strains were isolated from blood cultures, and none was reported from valvular tissue. In our case prior antibiotic treatment may have rendered the cultures sterile, and the culture vials (BACTEC, reference 4402192; Becton Dickinson) used in our laboratory were not supplemented with 0.01% l-cysteine hydrochloride impairing the growth of Granulicatella (13). Our patient is the first case in whom G. elegans was directly identified on the removed valve. We believe that culture of resected endocardial specimens is not sufficient to verify the presence of microorganisms and that broad-range PCR of the resected material should be performed to establish the diagnosis of IE postsurgically and to identify the tissue-causative agent, if the culture is negative (7, 11). G. elegans should be considered a new agent of IE.
REFERENCES
- 1.Bouvet, A., F. Grimont, and P. A. Grimont. 1989. Streptococcus defectivus sp. nov. and Streptococcus adjacens sp. nov., nutritionally variant streptococci from human clinical specimens. Int. J. Syst. Bacteriol. 39:290-294. [DOI] [PubMed] [Google Scholar]
- 2.Brouqui, P., and D. Raoult. 2001. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 14:177-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen, J. J., and R. R. Facklam. 2001. Granulicatella and Abiotrophia species from human clinical specimens. J. Clin. Microbiol. 39:3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., and P. A. Lawson. 2000. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella comb. nov. and Granulicatella balaenopterae comb. nov. Int J. Syst. E vol. Microbiol. 50:365-369. [DOI] [PubMed] [Google Scholar]
- 5.Fournier, P. E., and D. Raoult. 1999. Nonculture laboratory methods for the diagnosis of infectious endocarditis. Curr. Infect. Dis. Rep. 1:136-141. [DOI] [PubMed] [Google Scholar]
- 6.Frenkel, A., and W. Hirsch. 1961. Spontaneous development of L. forms of streptococci requiring secretions of other bacteria or sulphydryl compounds for normal growth. Nature 191:728-730. [DOI] [PubMed] [Google Scholar]
- 7.Goldenberger, D., A. Kunzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 3:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanamoto, T., S. Sato, and M. Inoue. 2000. Genetic heterogeneities and phenotypic characteristics of strains of the genus Abiotrophia and proposal of Abiotrophia para-adiacens sp. nov. J. Clin. Microbiol. 38:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura, Y., X.-G. Hou, F. Sultana, S. Liu, H. Yamamoto, and T. Ezaki. 1995. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int. J. Syst. Bacteriol. 45:798-803. [DOI] [PubMed] [Google Scholar]
- 10.La Scola, B., G. Michel, and D. Raoult. 1997. Use of amplification and sequencing of the 16S rRNA gene to diagnose Mycoplasma pneumoniae osteomyelitis in a patient with hypogammaglobulinemia. Clin. Infect. Dis. 24:1161-1163. [DOI] [PubMed] [Google Scholar]
- 11.Millar, B. C., J. E. Moore, P. W. G. Mallon, X. Jiru, M. J. Crowe, R. B. McClurg, D. Raoult, J. A. P. Earle, R. Hone, and P. G. Murphy. 2001. Molecular diagnosis of infective endocarditis—a new Duke's criterion? Scand. J. Infect. Dis. 33:673-680. [DOI] [PubMed] [Google Scholar]
- 12.Roggenkamp, A., L. Leitritz, K. Baus, E. Falsen, and J. Heeseman. 1998. PCR to detection and identification of Abiotrophia spp. J. Clin. Microbiol. 36:2844-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roggenkamp, A., M. Abele-Horn, K. H. Trebesius, U. Tretter, I. B. Autenrieth, and J. Heesemann. 1998. Abiotrophia elegans sp. nov., a possible pathogen in patient with culture-negative endocarditis. J. Clin. Microbiol. 36:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]