Abstract
The interpretation of molecular epidemiologic data of Mycobacterium tuberculosis infection is dependent on the understanding of the stability and evolutionary characteristics of the DNA fingerprinting marker used to classify clinical isolates. This study investigated the stability of the IS6110 banding pattern in serial tuberculosis isolates collected from patients resident in an area with a high incidence of tuberculosis. Evolutionary changes were observed in 4% of the strains, and a half-life (t1/2) of 8.74 years was calculated, assuming a constant rate of change over time. This rate may be composed of a high rate of change seen during the early disease phase (t1/2 = 0.57 years) and a low rate of change seen in the late disease phase (t1/2 = 10.69 years). The early rate probably reflects change occurring during active growth prior to therapy, while the low late rate may reflect change occurring during or after treatment. We demonstrate that the calculation of these rates is strongly influenced by the time interval between onset of disease and sputum sampling. These calculations are further complicated by partial replacement of the original strain population, resulting in the sporadic appearance of clonal variants in sputum specimens. Therefore, the true extent of genetic diversity may be underestimated within each host, thereby influencing molecular epidemiological data used to establish transmission chains.
Molecular epidemiology needs to differentiate clinical isolates of pathogenic organisms as being the same or different. This differentiation relies on the evolutionary process generating genotypic diversity among isolates. In general, the observation of a number of isolates sharing identical patterns is used to infer infection from a common source. In contrast, isolates with unrelated genotypes are thought to represent infection with different organisms, and therefore from independent sources.
The molecular epidemiology of Mycobacterium tuberculosis is most frequently based on the restriction fragment length polymorphism (RFLP) banding patterns generated by Southern hybridization with the probe IS6110 (10, 12). In community settings isolates with identical IS6110 genotypes are grouped together into clusters and are thought to represent recent epidemiological events (5), while isolates with IS6110 banding patterns unrelated to any others in the database (isolates with unique banding patterns) are thought to reflect reactivation of latent infection (1, 9). This methodology has been used in numerous settings to quantify the relative proportion of recent epidemiological events and thereby estimate the extent of recently transmitted disease (1, 9, 14).
In order to estimate the stability of the IS6110 RFLP patterns, studies have examined serial isolates collected from patients with persistent disease (3, 6, 15) and have demonstrated that the IS6110 banding pattern may change over time in a subset of these patients. When survival analysis was applied to the RFLP data collected from patients with persistent disease in The Netherlands, De Boer et al. (3) calculated that the half-life of the IS6110 banding pattern was on the order of 3.2 years. A similar rate was calculated for isolates originating from San Francisco, Calif., although this was restricted to isolates obtained at least 90 days apart, to exclude the possibility of variable patterns representing coinfection rather than change over time (3). In both data sets, the estimated rate of IS6110 pattern change was felt to be of the right magnitude to allow the use of IS6110 as a marker for molecular epidemiological studies (3).
However, the calculated rate of change may not be uniform in two groups of patients: those with multiple isolates at the time of diagnosis and those in whom a new pattern develops during or after treatment. In this study we have investigated the stability of the IS6110 banding pattern in a group of patients for whom more than one isolate of M. tuberculosis was collected during the course of disease or during retreatment. The results are discussed in the context of the applicability of calculating a rate of change in these patient groups and how such change may influence molecular epidemiological calculations.
MATERIALS AND METHODS
Study setting.
Between January 1992 and December 1998 M. tuberculosis isolates were collected from patients attending the healthcare clinics within adjacent suburbs in Cape Town, South Africa (2). This community is experiencing an extremely high incidence of tuberculosis, with approximately 250 new bacteriologically confirmed adult cases per 100,000 members of the population per year (11). Before 1995 the National Tuberculosis Program focused on sputum cultures for the diagnosis of tuberculosis, with follow-up sputum samples requested by the attending physician on clinical grounds. All sputum samples collected during this period that were positive and negative for Ziehl-Neelsen stain were included in the study. After 1995 the South African National Tuberculosis Program has been conducted according to the World Health Organization directly observed therapy, short course, strategy, with sputum taken for Ziehl-Neelsen smear at presentation for diagnosis, at 2 months for sputum conversion, and again at 6 months after initiation of therapy to monitor response to therapy (additional sputum specimens may be requested by the attending physician on clinical grounds). As part of the research project, all Ziehl-Neelsen-positive and -negative samples are sent for culture. Clinical information for each patient was recorded at the time of diagnosis, and these data were stored in a Microsoft Access database. The restricted sampling of sputum specimens during the course of disease in the different patients may influence rate calculations.
RFLP generation and GelCompar analysis.
The entire culture representing all the possible clonal variants present in each isolate was used for the DNA isolation, and this DNA was genotypically classified according to the internationally standardized DNA fingerprinting protocol, using the IS-3′ probe (10, 12). The Southern blot autoradiographs were normalized and the IS-3′ bands were assigned using GelCompar software (version 4.1). The assignments were visually checked by two independent persons, and only bands with an intensity of >20% of the other bands were scored as representing IS6110-mediated evolutionary events (4). Replicative transposition was identified when the evolved strain showed an additional IS6110 hybridizing band, while deletion of an IS6110 element was identified when an IS6110 hybridizing band was absent from the evolved strain. Variation in the electrophoretic mobility of an IS6110 hybridizing band was classified as a band shift representing a mutational event in the chromosomal domain flanking the IS6110 element. Banding pattern changes suspected of being the result of partial digestion of methylated restriction sites (13) were excluded.
Study population.
Patients with persistent disease from whom DNA fingerprints from more than one isolate were available were selected from the complete data set. This excluded isolates for which laboratory error (11), contamination, or loss of viability had been noted. Patients who had been assigned as being reinfected with a genotypically unrelated strain were treated as having two separate cases of disease if serial isolates were available for both infections. On average, three isolates were collected from each case of disease, and this was independent of the tuberculosis control program at the time of sample collection. Serial isolates collected from patients who presented with a genotypically identical or related strain after therapy were treated as a having a single case of disease. Selected patients were divided into two categories: (i) the group of patients infected with a strain which remained genotypically identical during the course of sampling was termed the “invariant group,” and (ii) the group of patients infected with a strain which changed during the course of sampling was termed the “variant group.” Change in the IS6110 banding pattern was identified by the appearance, disappearance, or electrophoretic shift of IS-3′ hybridizing elements. To avoid a possible bias in the variant patient group, variant strains with more than four changes in the IS6110 banding pattern were considered to reflect reinfection rather than evolution and were excluded from all calculations. In addition, if any variant strain (independent of the number of changes in the IS6110 banding pattern) was found to be present in the community prior to appearance in the patient, the patient was excluded from the study. In such a case, the patient may have been reinfected from a community contact. This stringent inclusion criteria excludes the possibility that the IS6110 banding pattern may revert to a previous evolutionary state or that identical banding patterns could have evolved convergently. It is envisaged that the above exclusion criteria will lead to an overestimate of the stability of the IS6110 banding pattern.
The sampling interval (in days) for patients infected with invariant strains was calculated from the date when the first isolate was collected to the date when the last isolate was collected in each patient. The sampling interval (in days) for patients infected with variant strains was calculated from the date when the first isolate was collected to the date when the first variant isolate was identified (3). The overall rate of change was calculated using survival analysis according to the method described by de Boer et al. (3). Similarly, the early rate of change was calculated using cases with interisolate intervals of ≤90 days, while the late rate of change was calculated using cases with interisolate intervals of >90 days.
Statistical analysis.
Fisher's exact test was used to test whether changes in the IS6110 banding pattern were associated with the number of IS6110 insertions present in the evolving strain. This test was also used to identify associations between previous therapy and the appearance of a variant strain. Chi-square analysis was used to identify differences between the study sample set and the sample set from The Netherlands (3).
RESULTS
Study population.
M. tuberculosis isolates were cultured from 954 patients (representing a recovery of approximately 70% of all culture-positive patients) attending the healthcare clinics in adjacent suburbs of Cape Town, South Africa (2, 14). Fifty of these patients (5.2%) were excluded from the study as their cultures were either contaminated or lost viability. Of the remaining patients, 901 (94.4%) had an M. tuberculosis isolate that could be genotypically classified by IS-3′ DNA fingerprinting (10, 12).
Patients with serial isolates of M. tuberculosis.
A total of 346 patients had two or more M. tuberculosis isolates, representing serial isolates from 351 active cases of tuberculosis. Analysis of the serial isolates by IS-3′ DNA fingerprinting identified 335 cases (95.4%) in which the serial M. tuberculosis isolates remained genotypically constant over the sampling period (range, 0 to 2,203 days), termed invariant strains (Table 1). Serial isolates from 16 cases showed an IS6110 banding pattern change over the sampling period, termed variant strains. Two of the patients infected with a variant strain were excluded, as the variant strain was found to have been present in the community prior to appearing in the patient, thereby suggesting possible exogenous reinfection from a community source. The remaining 14 variant cases were sampled over a period of 0 to 1,141 days (Table 1). Comparing patients with invariant strains to those with variant strains did not reveal any differences in the demographic variables. However, a previous history of disease was weakly associated with the appearance of variant strains (Fisher's exact test, P = 0.045) (Table 1), which may suggest that the evolution of clonal variants occurred either during the latent disease phase after the previous infection or during the reactivation process.
TABLE 1.
Demographics of patients from whom serial isolates of M. tuberculosis were collected
| Demographic parameter | Value for M. tuberculosis group
|
|
|---|---|---|
| Variant | Invariant | |
| Total no. of patients | 14 | 335 |
| No. (%) of male patients | 7 (50) | 187 (55.8) |
| Mean age (yr) | 35.6 | 34.1 |
| No. (%) of patients with: | ||
| Previous episode of TB | 9 (64)a | 120 (35.8) |
| Pulmonary TB | 14 (93.3) | 326 (97.3) |
| Sampling range (days) | 0-1,141 | 0-2,203 |
| No. of strains with: | ||
| Low IS6110 copy no. | 0b | 61 |
| High IS6110 copy no. | 14b | 274 |
P = 0.045 (Fisher's exact test for comparison with value from the invariant group).\
P = 0.142 (Fisher's exact test for comparison with value from the invariant group).
The proportion of variant strains identified (n = 14 [4%]) is similar to that reported for isolates collected in The Netherlands (4.6%) (3), although this is lower than the proportion reported in Germany (9%) (6) and in the restricted analysis performed in San Francisco (24.5%) (15). Change in the IS6110 banding pattern was specifically associated with strains with more than five hybridizing elements; however, because of the low number of observations, this was not statistically significant at the 95% confidence interval (95% CI) (Fisher's exact test, P = 0.142) (Table 1).
Analysis of the RFLP data of the variant isolates showed that in 10 of 14 cases (71.4%) the IS6110 banding patterns changed by replicative transposition, while in 2 of 14 cases (14.3%) a hybridizing band was deleted. In 2 of 14 cases (14.3%), an electrophoretic shift was observed, suggestive of a chromosomal mutation. For four patients the variant strain disappeared from subsequent serial isolates, demonstrating incomplete clonal replacement. This suggests that two clonal variant populations may be present in the patient and that a single sputum specimen may not reflect the true genetic diversity of the bacterial population in the host.
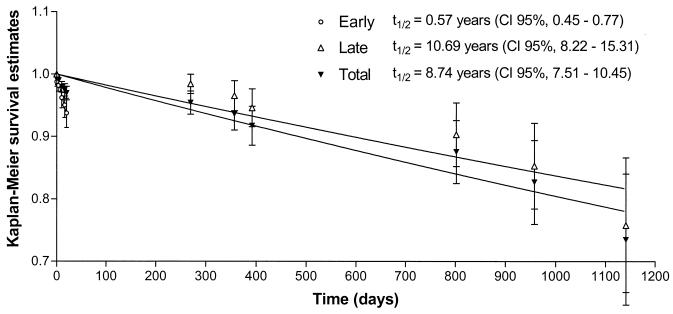
Survival analysis (3) of these data estimated the overall half-life of the IS6110 banding pattern in our serial isolates to be 8.74 years (95% CI, 7.51 to 10.45 years) (Fig. 1). However, the survival plot suggests the presence of two distinct rates of change (Fig. 1). In 8 of the 14 patients (57%) both the initial and variant isolate were sampled within the first 20 days (median of 10 days), suggesting that the banding pattern change occurred early in the disease process, during the latent phase of a previous infection, or during reactivation. This was followed by only partial clonal replacement at the time of diagnosis or shortly thereafter. In the remaining six patients (43%), variant isolates were only sampled after >250 days, suggesting that these isolates evolved late in the disease phase. Since all strains sampled between 20 days and 250 days were invariant, this provides further evidence for the argument that these late variant strains evolve over the course of time, rather than being due to under-sampling at an earlier point. Taking into account these two populations of observations, survival analysis estimated a half-life of 0.57 years (95% CI, 0.45 to 0.77 years) for the early group and 10.69 years (95% CI, 8.22 to 15.31 years) for the late group (Fig. 1).
FIG. 1.
Survival analysis of IS6110 fingerprint patterns using Kaplan-Meier estimates. The survival function is exp(−K × time), and the values of K for the early, late, and total variant groups are −0.003333, −0.0001776, and −0.0002173, respectively. t1/2, half-life.
DISCUSSION
The use of RFLP data to infer epidemiologic associations is dependent on an understanding of the marker system, including the sources and frequency of variability in that marker. To date, studies to determine the frequency of detection of variant strains have used different inclusion criteria and assumptions in order to provide an estimate on the “molecular clock” of RFLP for M. tuberculosis. The rate of change needs to be high enough to ensure that most cases not linked through recent transmission have different fingerprints but low enough to ensure that most cases linked through recent transmission have identical fingerprints. A half-life on the order of 2 to 5 years, as observed in San Francisco and The Netherlands, suggests that the IS6110 RFLP pattern changes rapidly enough but not too rapidly for studying ongoing transmission (3). A half-life on the order of 8 years, as observed in the present study, suggests that recent transmission may be overestimated by RFLP typing using IS6110.
In this study, the proportion of IS6110 banding pattern changes identified in serial isolates collected from patients with persistent disease was comparable to those reported in The Netherlands (3) but lower than those reported in Germany (6) and San Francisco (15). The comparable proportion of variant isolates seen in this study and the study from The Netherlands suggests that the rate of change is independent of the incidence of disease. However, in keeping with the observations of other groups, the ability to observe change appears to depend on the number of IS6110 elements present in the precursor strain, as strains with fewer than five IS6110 elements were genotypically invariant.
On further analysis, the emergence of these variant strains does not appear to be uniform, suggesting the presence of two distinct populations. The first type of change would occur early in the disease process, prior to antituberculosis treatment. This change might then represent evolution influenced by active growth or adaptation to the new host environment. It is also possible that change was induced in certain previously treated patients either during the latent growth phase or during the subsequent reactivation process. The second type of change would occur in the late disease phase or after treatment. This lower rate of change may be a consequence of a lower growth rate of the bacilli. Similar results were seen in data from The Netherlands (3). Although we have calculated an early rate, it is highly probable that this rate will be an underestimate of the stability of the banding pattern, as most of the IS6110 band changes will have occurred prior to the sampling of the first isolate. This notion is supported by the low in vivo growth rate of M. tuberculosis, requiring extended periods before strain population replacement would be observed. Furthermore, if there are long delays in the period between onset of early active disease and the first collection of sputa, then it is possible that newly evolved strains may be missed and thereby excluded from the rate calculations.
Due to the uncertainty of when the early evolutionary events occurred, we attempted to calculate a rate of change for variant isolates appearing after 90 days (15). The rate estimate of 10.69 years for variant isolates appearing after 90 days was significantly lower than the 2 years calculated from the San Francisco data (3). Review of the different rate calculation methods (3, 8) shows that these methods assume that the rate of appearance of variant strains is constant over time. If this assumption is incorrect, as is suggested by the present study, then the rate calculated by these methods (3, 8) will be sensitive to the proportion of (invariant) cases with extended interisolate time intervals arising due to either noncompliance, treatment failure, or treatment interruption. Comparison of our data with those from The Netherlands shows that there is a significant difference in the proportion of patients with invariant isolates after 210 days (this study, 21.2%; study from The Netherlands, 6.6%; P < 0.0001). This implies that if treatment efficacy is higher, the proportion of serial isolates with long intervals will be smaller, which may result in a higher rate of change estimate. Consequently, the accuracy of rate calculations is limited by the local epidemiology, and therefore rate estimates will be proportional to the effectiveness of the tuberculosis control program.
The epidemiological importance of calculating an evolutionary rate for isolates collected from patients with persistent disease is unclear, given that change is observed only in a very small and unique subset of the patients analyzed (3, 6, 15). Therefore, these data cannot be easily extrapolated to the overall study population. Furthermore, the true extent of genetic diversity within a study population may be masked by incomplete clonal replacement at the time of sputum collection, as extremely low numbers of clonal variants in the sputum specimens will not be detected due to the limited sensitivity of the DNA fingerprinting method. Therefore, strain variants will not be identified for patients from whom inadequate numbers of sputum specimens have been collected. Accordingly, it may be difficult to identify chains of transmission based only on genotypic identity (clustering of identical IS6110 banding patterns).
Although this study has highlighted the limitations of this sample set, we conclude that this patient set may provide information on how growth rate and/or antituberculosis therapy could influence the IS6110-mediated evolutionary rate. Furthermore, we do not exclude the possibility that change in the IS6110 banding pattern may be more frequent in other patient groups. To gain a greater insight into the rate of IS6110 change it will be essential to determine how transmission influences the evolutionary process (7).
Acknowledgments
This work was made possible by grants from the GlaxoSmithKline Action TB Initiative, the Sequella Global Tuberculosis Foundation, and the National Research Foundation (THRP).
E. Engelke, S. Carlini, and M. De Kock are thanked for their technical assistance.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Beyers, N., R. P. Gie, H. L. Zietsman, M. Kunneke, J. Hauman, M. Tatley, and P. R. Donald. 1996. The use of a geographical information system (GIS) to evaluate the distribution of tuberculosis in a high-incidence community. S. Afr. Med. J. 86:40-41, 44. [PubMed] [Google Scholar]
- 3.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, A. S., K. Kremer, M. W. Borgdorff, P. E. de Haas, H. F. Heersma, and D. van Soolingen. 2000. Genetic heterogeneity in Mycobacterium tuberculosis isolates reflected in IS6110 restriction fragment length polymorphism patterns as low-intensity bands. J. Clin. Microbiol. 38:4478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glynn, J. R., J. Bauer, A. S. de Boer, M. W. Borgdorff, P. E. Fine, P. Godfrey-Faussett, E. Vynnycky, et al. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 3:1055-1060. [PubMed] [Google Scholar]
- 6.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemann, S., S. Rüsch-Gerdes, E. Richter, H. Thielen, H. Heykes-Uden, and R. Diel. 2000. Stability of IS6110 restriction fragment length polymorphism patterns of Mycobacterium tuberculosis strains in actual chains of transmission. J. Clin. Microbiol. 38:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salamon, H., M. A. Behr, J. T. Rhee, and P. M. Small. 2000. Genetic distances for the study of infectious disease epidemiology. Am. J. Epidemiol. 151:324-334. [DOI] [PubMed] [Google Scholar]
- 9.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 10.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 12.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 13.van Soolingen, D., P. E. de Haas, R. M. Blumenthal, K. Kremer, M. Sluijter, J. E. Pijnenburg, L. M. Schouls, J. E. Thole, M. W. Dessens-Kroon, J. D. van Embden, and P. W. Hermans. 1996. Host-mediated modification of PvuII restriction in Mycobacterium tuberculosis. J. Bacteriol. 178:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren, R., J. Hauman, N. Beyers, M. Richardson, H. S. Schaaf, P. Donald, and P. van Helden. 1996. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S. Afr. Med. J. 86:45-49. [PubMed] [Google Scholar]
- 15.Yeh, R. W., A. Ponce de Leon, C. B. Agasino, J. A. Hahn, C. L. Daley, P. C. Hopewell, and P. M. Small. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J. Infect. Dis. 177:1107-1111. [DOI] [PubMed] [Google Scholar]