Abstract
Five pigmented isolates of Mycobacterium avium subsp. paratuberculosis were examined by pulsed-field gel electrophoresis (PFGE), IS900 restriction fragment length polymorphism (IS900-RFLP), and IS1311 polymorphism analysis using PCR. All of the pigmented isolates exhibited one of three distinct PFGE profiles with SnaBI, designated 9, 10, and 11, and with SpeI, designated 7, 8, and 9, which generated three multiplex profiles designated [9-7], [10-8], and [11-9]. All of the pigmented isolates had the same IS900-RFLP BstEII and PvuII profiles. The IS900-RFLP BstEII profile was new, but the IS900-RFLP PvuII profile corresponded to PvuII type 6 of a sheep strain described by Cousins and colleagues (D. V. Cousins, S. N. Williams, A. Hope, and G. J. Eamens, Aust. Vet. J. 78:184-190, 2000). IS1311-PCR analysis typed all of the pigmented isolates as sheep (S) strains. The genetic relationship between pigmented and nonpigmented isolates was investigated by using multiplex PFGE data from the analysis of both the 5 pigmented isolates and 88 nonpigmented isolates of M. avium subsp. paratuberculosis from a variety of host species and geographic locations. It was possible to classify the isolates into two distinct types designated type I, comprising the pigmented isolates, and type II, comprising the nonpigmented isolates, which exhibit a very broad host range.
Mycobacterium avium subsp. paratuberculosis is the etiological agent of paratuberculosis (Johne's disease), a fatal chronic granulomatous enteritis affecting principally ruminants. The organism is distinguished from other subspecies of M. avium by its mycobactin requirement for in vitro growth (22) and by the presence of the insertion sequence IS900 (9). Molecular typing procedures such as pulsed-field gel electrophoresis (PFGE) and restriction fragment length polymorphism (RFLP) coupled with hybridization to IS900 (IS900-RFLP) have shown that in comparison with other pathogens, there is relatively little genetic variability within this subspecies. However, there is a small subset of isolates distinguished by their prominent yellow or orange pigment that have not been characterized fully.
Pigmented M. avium subsp. paratuberculosis has been isolated in cases of lepromatous ovine paratuberculosis (18-20). In such cases, the affected intestine appears yellow or orange, probably due to the very large number of organisms present in the lesions. The organisms are difficult to culture, and few isolates have been maintained in collections. The pigmentation appears to be an inherent characteristic, as it is present at all stages of growth and is not altered by animal passage. The pigmented strains appear to have a host preference for sheep, although they can produce disease in cattle following experimental infection (21) and there has been one report of a naturally occurring bovine case (24). The purpose of this study was to analyze available pigmented isolates and investigate their genetic relatedness to other, nonpigmented M. avium subsp. paratuberculosis isolates.
MATERIALS AND METHODS
M. avium subsp. paratuberculosis isolates.
Five pigmented and 88 nonpigmented strains of M. avium subsp. paratuberculosis were analyzed in this study. Four of the pigmented strains were isolated at the Moredun Research Institute, Penicuik, Scotland, and Finn Saxegaard (National Veterinary Institute, Oslo, Norway) kindly provided pigmented strain 21P from the Faroe Islands. Nonpigmented isolates were obtained from the Veterinary Sciences Division of the Scottish Agricultural College, Perth, Scotland, the Veterinary Laboratories Agency, Starcross, England, and Queens University of Belfast, Belfast, Northern Ireland. M. avium subsp. paratuberculosis reference strains ATCC 19698 and NCTC 8578 were also included in the study. The nonpigmented isolates were obtained from a variety of tissues or fecal samples from infected animals, and 13 bovine isolates were cultured from milk samples as part of a survey funded by the Food Standards Agency (B08001). Nonpigmented isolates originated from different host species including cattle, sheep, goat, and deer; they also included isolates collected in a previous study by Beard et al. (2) from a number of wildlife species including rabbit (Oryctolagus cuniculus), hare (Lepus europaeus), fox (Vulpes vulpes), stoat (Mustela erminea), weasel (Mustela nivalis), badger (Meles meles), rat (Rattus norvegicus), wood mouse (Apodemus sylvaticus), rook (Corvus frugilegus), and crow (Corvus corone). The nonpigmented isolates were from different geographical areas within the United Kingdom: seven regions in Scotland, seven counties in England, and Northern Ireland (five isolates).
All isolates were propagated on slopes of Middlebrook 7H11 agar medium supplemented with 20% (vol/vol) heat-inactivated newborn calf serum, 2.5% (vol/vol) glycerol, 2 mM asparagine, 10% (vol/vol) Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment medium (Becton Dickinson, Oxford, Oxfordshire, United Kingdom), 2 Selectatabs (code MS 24; MAST Laboratories Ltd., Merseyside, United Kingdom) per liter, and 2 μg of mycobactin J (Allied Monitor, Fayette, Mo.) ml−1 for 8 to 10 weeks for nonpigmented isolates and as long as 6 months for pigmented isolates at 37°C. All isolates examined were fresh or low-passage-number isolates. Isolates were stored as glycerol stocks at −70°C.
PCR analysis.
All M. avium subsp. paratuberculosis isolates were screened for the presence of IS900 and IS1311 insertion sequences. Briefly, a single bacterial colony was suspended in 200 μl of sterile distilled water. Mycobacteria were lysed by using a Hybaid (Ashford, Middlesex, United Kingdom) RiboLyser at 5.5 m s−1 for 20 s, with cooling on ice before and after treatment. DNA was extracted by using guanidine hydrochloride (4) and was analyzed by PCR. Alternatively, DNA was prepared in low-melting-point agarose plugs as described below. Amplified products were detected on agarose gels. PCR analysis for IS900 was carried out by using the primers and PCR conditions described by Sanderson et al. (17). The IS900 PCR product was verified by hybridization to a 2,4-dinitrophenyl-labeled oligonucleotide probe complementary to the amplified sequence (2) and by restriction analysis with AlwI as reported by Cousins et al. (6). PCR analysis for IS1311 and restriction analysis to discriminate between other M. avium subspecies and M. avium subsp. paratuberculosis and between cattle and sheep strains were performed according to the work of Marsh et al. (12).
IS900 sequencing.
IS900 was amplified from pigmented strain M189 as described above. The PCR product was excised from the agarose gel and extracted by using AgarACE (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. The DNA was cloned into pGEM-T (Promega) and was sequenced on an ABI 377 sequencer. Sequence analysis was performed by using the software package Vector NTI (InforMax Inc., Oxford, United Kingdom).
PFGE analysis.
DNA for PFGE analysis was prepared from stirred broth cultures of M. avium subsp. paratuberculosis isolates as described previously (10). Briefly, 10 ml of Middlebrook 7H9 broth (supplemented with 0.4% [wt/vol] Tween 80, 10% [vol/vol] OADC, and 2 μg of mycobactin J ml−1) was inoculated with a single colony of M. avium subsp. paratuberculosis and incubated at 37°C with continuous stirring to prevent clumping of cells. When the cell density was at least approximately McFarland standard 2, cell suspensions were quantified by optical density by using a Densimat (BioMerieux, Lyon, France) and then harvested by centrifugation, washed, and resuspended in modified spheroplasting buffer. Bacterial suspensions (6 × 109cells ml−1) were mixed with an equal volume of 1.5% (wt/vol) low-melting-point agarose and poured into plug molds. Plugs were incubated in Tris-EDTA (pH 8) containing 1 to 2 mg of lysozyme ml−1 for at least 18 h at 37°C and then in 500 mM EDTA containing 1%(wt/vol) sarcosine and 1 to 2 mg of proteinase K ml−1 for 7 days at 55°C.
Slices (inserts) 3 to 5 mm thick were cut from the plugs, washed six times in 5 ml of Tris-EDTA (pH 8), preequilibrated in commercial restriction buffer containing bovine serum albumin, and then incubated with a restriction reaction mixture containing 10 U of restriction endonuclease ml−1. After overnight incubation, fresh restriction reaction mixture was added and incubation was continued for at least another 3 h to ensure complete digestion of the sample. Restricted samples were loaded onto a 1% (wt/vol) agarose gel and electrophoresed in 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA). SnaBI and SpeI restriction enzymes were purchased from New England Biolabs (Hitchin, Hertfordshire, United Kingdom).
Electrophoresis was conducted using a CHEF Mapper (Bio-Rad, Hemel Hempstead, Hertfordshire, Hertfordshire, United Kingdom). Parameters for electrophoresis were as follows. For SnaBI restriction fragments, initial and final switch times were 6.75 and 26.29 s, respectively, and with switch times ramped linearly; the gradient was 6 V cm−1; the included angle was 120o; and electrophoresis was carried out at 14°C for 40 h. For SpeI restriction fragments, initial and final switch times were 2.16 and 35.38 s, respectively, and with switch times ramped linearly; the gradient was 6 V cm−1; the included angle was 120o; and electrophoresis was carried out at 14°C for 23 h. Lambda midrange markers (catalog no. 355-2; New England Biolabs) were loaded onto gels as molecular size standards.
Nomenclature.
The different PFGE profiles for each enzyme were assigned numbers, with the ATCC 19698 type strain designated profile 1 in each case. This system was used to distinguish the nomenclature for PFGE from the alphabetical and numerical system used for IS900-RFLP (14). The different SnaBI-SpeI multiplex PFGE profiles have been expressed as the SnaBI and SpeI profile numbers joined by a hyphen and enclosed in square brackets. For example, the multiplex PFGE profile of an isolate that has a SnaBI profile of 2 and a SpeI profile of 3 is expressed as [2-3]. This system was chosen because it gives some indication of the genetic relationship between isolates, and adding new profiles is easy. A database of the PFGE profiles is available at www.mri.sari.ac.uk/Bacteriology/PFGE-mycobacteria.
Phylogenetic analysis.
Electronic images of PFGE profiles were initially analyzed by using the computer software package Imagemaster, version 3.0 (Amersham Pharmacia Biotech). Phylogenetic analysis was carried out on the PFGE data from this study, encompassing a total of 16 multiplex PFGE profiles. The PFGE data from the SpeI and SnaBI restriction enzyme digestions were combined to form a matrix of the 16 multiplex PFGE profiles and the presence or absence of 84 bands. The PHYLIP Phylogeny Inference Package version 3.6 2001 was used for the phylogenetic analysis, and the data were analyzed by two approaches; a distance-based approach and the maximum parsimony approach. Differences in band presence or absence are due to nucleotide substitution or processes that result in length differences (e.g., insertion, deletion, or duplication events) in the nucleotide sequences. If nucleotide substitution is the only evolutionary process acting, it is possible to infer changes at the nucleotide level from the band presence or absence data and to construct phylogenetic trees with branch lengths proportional to the genetic change (nucleotide substitutions per position). Modelling both nucleotide substitution and insertion or deletion events for PFGE data is not currently possible, so a simple parsimony analysis is a reasonable approach. Both approaches were used, since the M. avium subsp. paratuberculosis genome is known to contain a number of insertion sequences.
(i) Distance-based approach.
Genetic distances were calculated by using the RESTDIST program (PHYLIP package, version 3.6) with plausible values for the nucleotide substitution process (transition/transition ratio = 2; gamma rate heterogeneity parameter alpha = 4) based on the Jin/Nei model. The distance matrix was then analyzed by the Fitch-Margoliash method (with the FITCH program in the PHYLIP package).
(ii) Maximum parsimony approach.
The data were analyzed by using the PHYLIP DOLLOP program (Dollo parsimony method for binary data). Statistical testing of the clusters within the phylogenetic tree for both distance-based and parsimony analysis was carried out by using bootstrapping with 100 trials.
IS900-RFLP analysis.
DNA for RFLP analysis was prepared in low-melting-point agarose plugs, washed, and treated with restriction endonucleases as described for PFGE analysis. DNA was restricted with BstEII (catalog no. R6641; Promega UK Ltd.) or PvuII (catalog no. 642 690; Roche) by using the conditions recommended by the manufacturer. Following digestion, inserts were melted in a water bath at 70°C for 5 min and then loaded onto a 0.8% (wt/vol) agarose gel. DNA fragments were separated by conventional gel electrophoresis in 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 20 V overnight. Lambda-HindIII and 1-kb DNA ladders (catalog no. G5711; Promega UK Ltd.) were used as molecular size markers.
After electrophoresis, the gel was treated with denaturation solution (1.5 M sodium chloride-0.5 M sodium hydroxide) for 1 h followed by neutralizing solution (1.5 M sodium chloride-0.5 M Tris-HCl adjusted to pH 7.5) for 1 h. All incubations were carried out at room temperature with constant gentle agitation. DNA fragments were transferred to Hybond-N nylon membranes (Amersham Pharmacia) by standard blotting techniques (11) and heat fixed onto the membranes for 2 h at 80°C. DNA was hybridized overnight at 65°C to a probe derived from IS900 and randomly labeled by using the Gene Images random prime-labeling module (catalog no. RPN 3540; Amersham Pharmacia). The membrane was washed once for 5 min in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate [pH 7])-0.1% (wt/vol) sodium dodecyl sulfate at 42 to 65°C, followed by three 5-min washes in 0.1× SSC-0.1% (wt/vol) sodium dodecyl sulfate at 65°C, and then wiped with a tissue soaked in 0.1× SSC at 65°C. Detection of the fluorescein-labeled probe was carried out by using the Gene Images CDP-Star detection module (catalog no. 3510; Amersham Pharmacia) according to the manufacturer's instructions.
RESULTS
All of the pigmented isolates analyzed in this study were obtained from lepromatous cases of ovine paratuberculosis, and the intestines were characteristically pigmented (Fig. 1). The organisms grew very slowly in culture and were acid fast as determined by Ziehl-Neelsen staining. Single colonies were smooth, convex, and glistening with a yellow pigment. By contrast, colonies of nonpigmented isolates of M. avium subsp. paratuberculosis were rough and had a creamy color. The pigmented organisms were coccobacillary as opposed to the bacillary nonpigmented isolates, as determined by light microscopy.
FIG. 1.
Opened segment of ileum from a case of pigmented ovine paratuberculosis.
All of the pigmented isolates and nonpigmented isolates of M. avium subsp. paratuberculosis analyzed in this study were found to be positive for IS900 by PCR. Since IS900-like sequences have been detected by PCR in a few other species of mycobacteria (6), the PCR products were verified as IS900 by restriction analysis and hybridization to a probe. In addition, the PCR product from pigmented isolate M189 was sequenced, and the sequence was found to be identical to that reported for M. avium subsp. paratuberculosis (GenBank accession number X16293). All isolates analyzed in this study possessed IS1311 sequences and were confirmed as M. avium subsp. paratuberculosis by restriction of the PCR product with MseI (26). Restriction of the IS1311 PCR product with HinfI detects a point mutation at base position 223 in C strains, isolated mainly from cattle, that is absent in S strains, isolated mainly from sheep. Restriction analysis of the isolates revealed that the pigmented isolates were of the S type and the nonpigmented isolates, including those isolated from sheep, were of the C type.
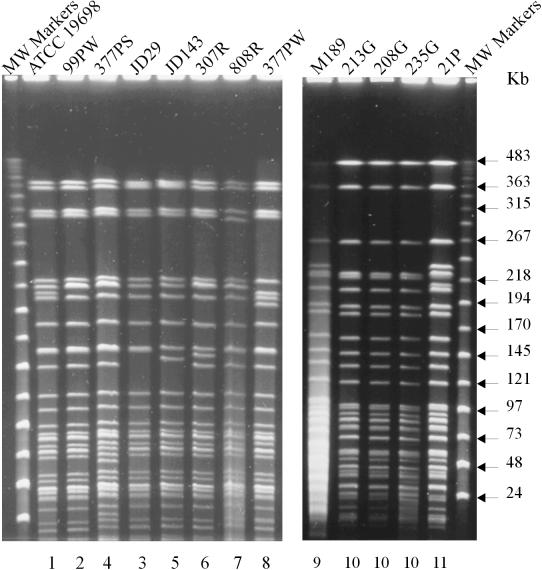
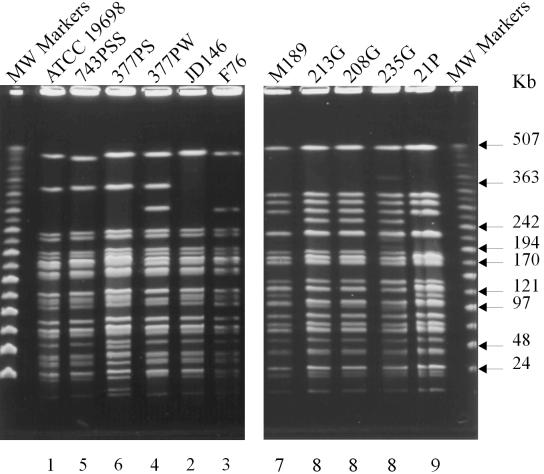
The polymorphisms that define all of the pulsed-field types are clearly visible under the specified running conditions. Combining the results of SnaBI and SpeI generates 16 multiplex PFGE profiles (Table 1). All of the isolates were analyzed by multiplex PFGE using the enzymes SnaBI and SpeI. Restriction of the pigmented isolates with these enzymes yielded profiles distinct from those of the nonpigmented isolates. Three distinct profiles, designated 9, 10, and 11, were identified with SnaBI (Fig. 2), and three distinct profiles, designated 7, 8, and 9, were identified with SpeI (Fig. 3), which generated three multiplex profiles designated [9-7], [10-8], and [11-9] (Table 1). A total of 88 nonpigmented isolates of M. avium subsp. paratuberculosis were analyzed by multiplex PFGE. By use of SnaBI, eight PFGE types were detected, which were designated 1 to 8, with ATCC 19698 designated as type 1. The PFGE profiles of eight representative strains are presented in Fig. 2. When the nonpigmented isolates were typed with SpeI, six PFGE profiles were detected. The PFGE profiles of six representative strains are presented in Fig. 3. Of the nonpigmented isolates, 18 were from an ovine host. Thirteen of these ovine isolates were found to have multiplex profile [2-1], two isolates had [5-2], and one isolate each had multiplex profile [2-2], [3-1], or [3-3].
TABLE 1.
PFGE and RFLP profiles of M. avium subsp. paratuberculosis reference isolates
| Isolatea | Host species | Geographic origin | Multiplex PFGE profile | BstEII RFLP profileb | PvuII RFLP profilec |
|---|---|---|---|---|---|
| ATCC 19698 | Bovine | Unknown | [1-1] | C1 | 1 |
| 99PW | Bovine | Staffordshire, England | [2-1] | C5 | Npad |
| JD146 | Ovine | Perth & Kinross, Scotland | [2-2] | C17 | 3 |
| JD29 | Ovine | Angus, Scotland | [3-1] | C17 | 3 |
| R197 | Leporine | Perth & Kinross, Scotland | [3-2] | C17 | 3 |
| F76 | Ovine | Perth & Kinross, Scotland | [3-3] | C17 | 3 |
| 377PS | Bovine | Shropshire, England | [4-6] | Nca | 1 |
| JD18 | Bovine | Aberdeenshire, Scotland | [5-1] | C17 | 3 |
| JD143 | Ovine | Perth & Kinross, Scotland | [5-2] | C17 | 3 |
| 307R | Bovine | Lancashire, England | [6-5] | C5 | Npa |
| 808R | Bovine | Northern Ireland | [7-1] | C5 | Npa |
| 743PSS | Bovine | Midlothian, Scotland | [7-5] | C1 | 1 |
| 377PW | Bovine | Shropshire, England | [8-4] | Ncb | 2 |
| M189 | Ovine | Midlothian, Scotland | [9-7] | Ncc | 6 |
| 213G, 208G, 235G | Ovine | Shetland, Scotland | [10-8] | Ncc | 6 |
| 21P | Ovine | Faroe Islands | [11-9] | Ncc | 6 |
FIG. 2.
PFGE profiles of M. avium subsp. paratuberculosis isolates generated with SnaBI. (Left) Profiles of nonpigmented isolates; (right) profiles of pigmented ovine isolates. Numbers above the lanes are designations of isolates, details for which are given in Table 1. Numbers below the lanes represent the different PFGE SnaBI reference types. PFGE running conditions are described in Materials and Methods.
FIG. 3.
PFGE profiles of M. avium subsp. paratuberculosis isolates generated with SpeI. (Left) Profiles of nonpigmented isolates; (right) profiles of pigmented ovine isolates. Numbers above the lanes are designations of isolates, details for which are given in Table 1. Numbers below the lanes represent the PFGE SpeI reference types. PFGE running conditions are described in Materials and Methods.
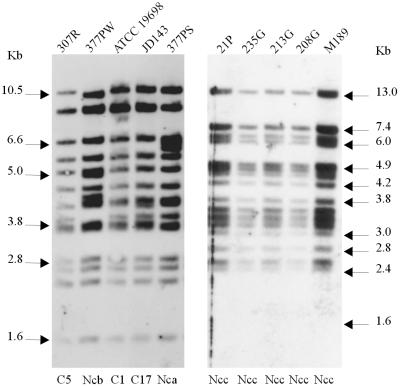
All of the pigmented isolates and one representative nonpigmented strain for each of the multiplex PFGE profiles were analyzed by IS900-PCR using BstEII and PvuII. All of the pigmented isolates had the same IS900-RFLP BstEII profile (Fig. 4). This profile appears to be similar to the IS900-RFLP BstEII S profiles previously described by Collins et al. (5), Whittington et al. (27), and Cousins et al. (7). The pigmented isolates had identical IS900-RFLP PvuII profiles (Fig. 5), which appeared to correspond to PvuII type 6 (described by Cousins et al. [7]). In contrast, nonpigmented isolates representative of the different multiplex PFGE profiles were found to have BstEII profiles corresponding to C1, C5, and C17 (defined by Pavlik et al. [15]) and two new profiles not previously published (Table 1) and PvuII profiles corresponding to types 1, 2, and 3 (defined by Whipple et al. [25]) and one new profile not previously described.
FIG. 4.
IS900-RFLP profiles of M. avium subsp. paratuberculosis isolates generated with BstEII. (Left) Profiles of nonpigmented isolates; (right) profiles of pigmented ovine isolates. Numbers above the lanes are designations of isolates, details for which are given in Table 1. Numbers below the lanes represent the IS900-RFLP types as defined by Pavlik et al. (15). Nca, Ncb, and Ncc, new profiles not previously published.
FIG. 5.
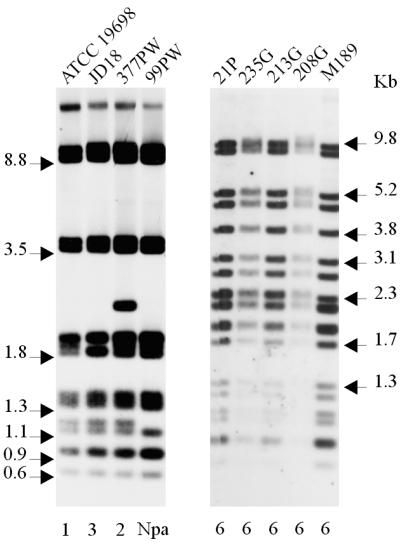
IS900-RFLP profiles of M. avium subsp. paratuberculosis isolates generated with PvuII. (Left) Profiles of nonpigmented isolates; (right) profiles of pigmented ovine isolates. Numbers above the lanes are designations of isolates, details for which are given in Table 1. Numbers below the lanes represent the IS900-RFLP types as defined by Whipple et al. (25).
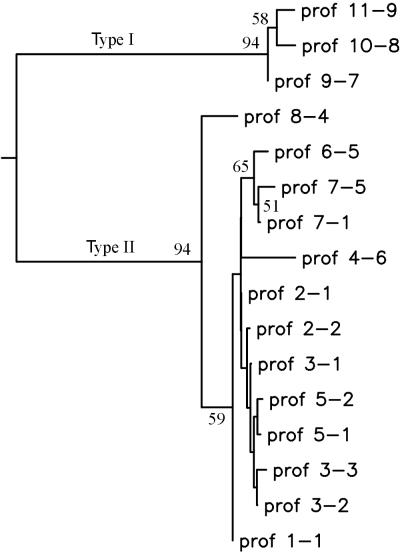
Both a distance-based approach and the maximum parsimony approach were used to estimate the genetic relatedness of pigmented to nonpigmented isolates. The results of the distance-based approach are given below and in Fig. 6. The consensus best tree and the bootstrap support values obtained by the parsimony approach were very similar to those obtained by the distance-based approach (data not shown).
FIG. 6.
Dendrogram of DNA convergence generated from the 16 multiplex PFGE profiles (prof) of pigmented and nonpigmented isolates of M. avium subsp. paratuberculosis. Bootstrap support values greater than 50% are shown, although only those greater than 70% are considered statistically significant.
The phylogenetic tree in Fig. 6 is resolved into two significant clusters: a minor cluster of 3 profiles (containing profiles [9-7], [10-8], and [11-9] representing the pigmented isolates) and a major cluster of 13 profiles (containing all the profiles of nonpigmented isolates, including bovine, ovine, cervine, caprine, and wildlife M. avium subsp. paratuberculosis isolates). Bootstrap support values greater than 50% are shown in the figure, although only those greater than 70% are considered statistically significant. The branching order within the clusters is not statistically significant.
If the difference between profiles is due only to nucleotide substitution events, then the branch lengths from the distance-based analysis give an idea of the divergence in the nucleotide sequences. The long branch between the two clusters is 0.027 nucleotide substitutions per position, representing approximately 2.7% change.
DISCUSSION
This is the first report of the molecular characterization of pigmented isolates of M. avium subsp. paratuberculosis and the most comprehensive multiplex PFGE study of M. avium subsp. paratuberculosis isolates to date. Molecular typing by PFGE, IS900-RFLP, and IS1311-PCR has revealed that these isolates are genetically closely related to the S strains described by Collins et al. (5), de Lisle et al. (8), Bauerfeind et al. (1), Cousins et al. (7), and Whittington et al. (27). M. avium subsp. paratuberculosis isolates analyzed by PFGE to date appear to segregate into two distinct clusters; cluster I encompasses the pigmented ovine isolates, and cluster II comprises isolates from a wider host range. Isolates within a cluster share many phenotypic and genetic characteristics but also show a small amount of genetic heterogeneity. This division into two clusters supports the earlier observation that two forms of ovine paratuberculosis exist in Britain, one caused by a strain of the organism prevalent in cattle and one by a slow-growing pigmented strain (19, 20). The data are also consistent with the division of isolates into two types of strains, S and C, as reported by Collins et al. (5) and Bauerfeind et al. (1). The nomenclature used for this strain classification is confusing and misleading, as it suggests that there is an exclusive correlation between strain type and host species of origin. While there may be circumstantial evidence for strong host preferences, there is no evidence for host-specific strains of M. avium subsp. paratuberculosis. In Australia and New Zealand, S strains are the predominant strains isolated from sheep (7, 27). However, in Europe, where paratuberculosis is endemic, C strains are more commonly isolated from sheep as well as from cattle and nonruminant hosts (13, 16). Furthermore, pigmented isolates can infect and produce disease in cattle (21, 24; B. Hanusson, personal communication), and S strains from the Faroe Islands, Iceland, and Australia have been isolated from cattle (28). Therefore, we propose that the two genetically distinct clusters of isolates be referred to as type I and type II. Type I comprises the slow-growing strains that appear to have a strong host preference for sheep and may be more virulent for sheep (18). These include the pigmented strains, ovine isolates from Iceland (28), Morocco (1), South Africa (1, 8), Australia (7, 27), and New Zealand (5), and a few Norwegian caprine strains (5, 8, 23). Type II includes the faster-growing strains commonly isolated from cattle and a broad host range including humans and a variety of wildlife species (2).
The geographic distribution of pigmented strains is intriguing. In Britain, cases of pigmented paratuberculosis have been reported in Scotland and England (18, 19). Based on records kept by the Scottish Agricultural College Veterinary Sciences Division in Scotland, between 1996 and 2000 approximately 19% of confirmed cases of ovine paratuberculosis were caused by pigmented isolates (A. Greig, personal communication). Pigmented ovine paratuberculosis appears to be more prevalent in southern Scotland. No cases of pigmented bovine paratuberculosis were reported during this period, and no type I nonpigmented strains have been isolated as far as is known. However, in Scotland most cases of paratuberculosis are diagnosed on the basis of serological testing or fecal smears, and cases of pigmented paratuberculosis are likely to be diagnosed only at a postmortem examination or at the abattoir. Elsewhere in Europe, pigmented ovine paratuberculosis has been reported in the Faroe Islands (F. Saxegaard, personal communication; B. Hanusson, personal communication) and Spain (R. A. Juste, personal communication). Outside Europe, there are no reported cases of pigmented paratuberculosis except in Morocco, where 3 of 56 ovine isolates were found to be pigmented (3). Despite the prevalence of nonpigmented type I strains in sheep in Australia and New Zealand, there has been no known case of pigmented paratuberculosis in these countries (D. Kennedy, personal communication; G. W. de Lisle, personal communication). Why the distribution of pigmented strains should be restricted to certain geographic regions is a matter for conjecture. There does not appear to be any correlation with breeds of sheep, and there are no known environmental factors that could explain the distribution.
This study has demonstrated that PFGE can detect more genetic diversity than IS900-RFLP alone. In the panel of reference isolates, 16 multiplex PFGE profiles versus 6 multiplex IS900-RFLP profiles were detected. Multiplex PFGE clearly has the ability to subdivide IS900-RFLP types. For example, multiplex PFGE subdivides BstEII profile C1 into two types ([1-1] and [7-5]), BstEII profile C5 into three types ([2-1], [6-5], and [7-1]), and BstEII profile C17 into six types ([2-2], [3-1], [3-2], [3-3], [5-1], and [5-2]). Although IS900-RFLP currently is the most widely used technique for discriminating strains of M. avium subsp. paratuberculosis, it has some important limitations. IS900-RFLP does not detect sufficient genetic diversity, and in many countries a single IS900-RFLP type predominates, limiting its use for epidemiological monitoring and surveillance. Furthermore, the discriminatory power of IS900-based typing techniques depends on the number of copies and insertion loci of IS900 in any one strain; such techniques, therefore, are inherently limited in their ability to detect genetic polymorphisms. Multiplex PFGE analysis explores the whole genome and may be more appropriate and successful for discriminating organisms with limited genetic heterogeneity. In this study we looked only at the IS900-RFLP types of the 16 PFGE reference strains. Testing a larger number of isolates of the same multiplex PFGE profile may identify more IS900-RFLP types. Combining both techniques may achieve even greater discrimination of M. avium subsp. paratuberculosis, which will greatly assist epidemiological studies on a national or global scale.
Acknowledgments
We thank A. Greig (Scottish Agricultural College, Veterinary Sciences Division), I. Grant (Queens University of Belfast), M. P. Cranwell (Veterinary Laboratories Agency, Starcross, England), and F. Saxegaard (National Veterinary Institute, Oslo, Norway) for provision of M. avium subsp. paratuberculosis isolates; Kathleen Connor, Amanda Pirie, and Karen Rudge for excellent technical assistance; and Brian Easter for photography (Moredun Research Institute).
This study was funded by the Scottish Executive Environment and Rural Affairs Department (SEERAD). L. de Juan is a Ph.D. student funded by the Ministerio de Ciencia y Tecnología of Spain.
REFERENCES
- 1.Bauerfeind, R., S. Benazzi, R. Weiss, T. Schliesser, H. Willems, and G. Baljer. 1996. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J. Clin. Microbiol. 34:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard, P. M., M. J. Daniels, D. Henderson, A. Pirie, K. Rudge, D. Buxton, S. Rhind, A. Greig, M. R. Hutchings, I. McKendrick, K. Stevenson, and J. M. Sharp. 2001. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 39:1517-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benazzi, S., M. el Hamidi, and T. Schliesser. 1996. Paratuberculosis in sheep flocks in Morocco: a serological, microscopical and cultural survey. Zentbl. Veterinarmed. B 43:213-219. [DOI] [PubMed] [Google Scholar]
- 4.Challans, J. A., K. Stevenson, H. W. Reid, and J. M. Sharp. 1994. A rapid method for the extraction and detection of Mycobacterium avium subspecies paratuberculosis from clinical specimens. Vet. Rec. 134:95-96. [DOI] [PubMed] [Google Scholar]
- 5.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 7.Cousins, D. V., S. N. Williams, A. Hope, and G. J. Eamens. 2000. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp. paratuberculosis using IS900 RFLP. Aust. Vet. J. 78:184-190. [DOI] [PubMed] [Google Scholar]
- 8.de Lisle, G. W., D. M. Collins, and H. F. Huchzermeyer. 1992. Characterization of ovine strains of Mycobacterium paratuberculosis by restriction endonuclease analysis and DNA hybridization. Onderstepoort J. Vet. Res. 59:163-165. [PubMed] [Google Scholar]
- 9.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes, V. M., K. Stevenson, and J. M. Sharp. 2001. Improved preparation of high molecular weight DNA for pulsed-field gel electrophoresis from mycobacteria. J. Microbiol. Methods 44:209-215. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Marsh, I., R. Whittington, and D. Cousins. 1999. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol. Cell. Probes 13:115-126. [DOI] [PubMed] [Google Scholar]
- 13.Pavlik, I., J. Bartl, L. Dvorska, P. Svastova, R. du Maine, M. Machackova, W. Yayo Ayele, and A. Horvathova. 2000. Epidemiology of paratuberculosis in wild ruminants studied by restriction fragment length polymorphism in the Czech Republic during the period 1995-1998. Vet. Microbiol. 77:231-251. [DOI] [PubMed] [Google Scholar]
- 14.Pavlik, I., L. Bejckova, M. Pavlas, Z. Rozsypalova, and S. Koskova. 1995. Characterization by restriction endonuclease analysis and DNA hybridization using IS900 of bovine, ovine, caprine and human dependent strains of Mycobacterium paratuberculosis isolated in various localities. Vet. Microbiol. 45:311-318. [DOI] [PubMed] [Google Scholar]
- 15.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 16.Pavlik, I., A. Horvathova, L. Dvorska, P. Svastova, R. du Maine, B. Fixa, and I. Rychlik. 1999. Homogeneity/heterogeneity of Mycobacterium avium subsp. paratuberculosis strains: correlation between RFLP-type and source (animal, environment, human), p. 321-329. In E. J. B. Manning and M. T. Collins (ed.), Proceedings of the Sixth International Colloquium on Paratuberculosis. International Association for Paratuberculosis, Inc., Madison, Wis.
- 17.Sanderson, J. D., M. T. Moss, M. L. Tizard, and J. Hermon-Taylor. 1992. Mycobacterium paratuberculosis DNA in Crohn's disease tissue. Gut 33:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamp, J. T., and J. A. Watt. 1954. Johne's disease in sheep. J. Comp. Pathol. 64:26-40. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, A. W. 1945. Ovine paratuberculosis (Johne's disease of sheep). J. Comp. Pathol. 55:41-44. [Google Scholar]
- 20.Taylor, A. W. 1951. Varieties of Mycobacterium johnei from sheep. J. Pathol. Bacteriol. 63:333-336. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, A. W. 1953. The experimental infection of cattle with varieties of Mycobacterium johnei isolated from sheep. J. Comp. Pathol. 63:368-373. [DOI] [PubMed] [Google Scholar]
- 22.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 23.Thoresen, O. F., and I. Olsaker. 1994. Distribution and hybridization patterns of the insertion element IS900 in clinical isolates of Mycobacterium paratuberculosis. Vet. Microbiol. 40:293-303. [DOI] [PubMed] [Google Scholar]
- 24.Watt, J. A. A. 1954. Johne's disease in a bovine associated with the pigmented strain of Mycobacterium johnei. Vet. Rec. 66:387. [Google Scholar]
- 25.Whipple, D., P. Kapke, and C. Vary. 1990. Identification of restriction fragment length polymorphisms in DNA from Mycobacterium paratuberculosis. J. Clin. Microbiol. 28:2561-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 27.Whittington, R. J., A. F. Hope, D. J. Marshall, C. A. Taragel, and I. Marsh. 2000. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS900 restriction fragment length polymorphism and IS1311 polymorphism analyses of isolates from animals and a human in Australia. J. Clin. Microbiol. 38:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whittington, R. J., C. A. Taragel, S. Ottaway, I. Marsh, J. Seaman, and V. Fridriksdottir. 2001. Molecular epidemiological confirmation and circumstances of occurrence of sheep (S) strains of Mycobacterium avium subsp. paratuberculosis in cases of paratuberculosis in cattle in Australia and sheep and cattle in Iceland. Vet. Microbiol. 79:311-322. [DOI] [PubMed] [Google Scholar]