Abstract
Background
In most organisms proper reductional chromosome segregation during meiosis I is strongly correlated with the presence of crossover recombination structures (chiasmata); recombination deficient mutants lack crossovers and suffer meiosis I nondisjunction. We report that these functions are separable in the fission yeast Schizosaccharomyces pombe.
Results
Intron mapping and expression studies confirmed that Rec12 is a member of the Spo11/Top6A topoisomerase family required for the formation of meiotic dsDNA breaks and recombination. rec12-117, rec12-D15 (null), and rec12-Y98F (active site) mutants lacked most crossover recombination and chromosomes segregated abnormally to generate aneuploid meiotic products. Since S. pombe contains only three chromosome pairs, many of those aneuploid products were viable. The types of aberrant chromosome segregation were inferred from the inheritance patterns of centromere linked markers in diploid meiotic products. The rec12-117 and rec12-D15 mutants manifest segregation errors during both meiosis I and meiosis II. Remarkably, the rec12-Y98F (active site) mutant exhibited essentially normal meiosis I segregation patterns, but still exhibited meiosis II segregation errors.
Conclusions
Rec12 is a 345 amino acid protein required for most crossover recombination and for chiasmatic segregation of chromosomes during meiosis I. Rec12 also participates in a backup distributive (achiasmatic) system of chromosome segregation during meiosis I. In addition, catalytically-active Rec12 mediates some signal that is required for faithful equational segregation of chromosomes during meiosis II.
Background
During meiosis homologous chromosomes replicate, pair to form a "bivalent," experience a high rate of recombination, and undergo two rounds of chromosome segregation to produce haploid meiotic products. In meiosis I (MI) homologous chromosomes segregate from their partners in a reductional division, and during meiosis II (MII) sister chromatids segregate from one-another in an equational division similar to mitosis.
Crossover recombination (reciprocal exchange) generates chiasmata, physical connections between homologs, which provide the primary mechanism to ensure proper segregation of homologs during MI [1]. In the absence of chiasmata (crossovers), homologous chromosomes experience nondisjunction and segregate randomly from their partners. However, backup achiasmatic or "distributive" mechanisms have been identified in some species [2-7]. These distributive segregation systems can partially or almost fully restore faithful segregation of chromosomes during MI in the absence of recombination.
The respective contributions of chiasmatic and achiasmatic (distributive) modes of chromosome segregation can be studied in organisms with naturally-occurring achiasmatic chromosomes [8], in cells harboring non-recombined artificial chromosomes [9], and in achiasmatic mutants [10]. Among achiasmatic mutants, those lacking the Spo11 (Rec12) protein are of particular interest because Spo11 has a key role in the initiation of recombination [11].
Meiotically induced, Spo11 (Rec12)-dependent double-strand DNA (dsDNA) breaks have been demonstrated in budding yeast and fission yeast [12-15]. Spo11 almost certainly catalyzes these breaks, as it shares high homology with the A subunit of Topoisomerase VI of Sulfolobus shibatae [16,17] and it becomes linked by a phosphotyrosine linkage to the 5' end of the cleaved DNA [18-21]. Spo11 homologs have been identified in a wide range of eukaryotes, and available evidence suggests that initiation of meiotic recombination via Spo11-dependent dsDNA break formation is conserved [22-25]. Our results with the fission yeast S. pombe fully support this view (see results and discussion).
Spo11 protein has potential functions in addition to catalyzing dsDNA breaks. It is dispensable for chromosome pairing and synapsis in some organisms, but is required in others [22,24-28]. Budding yeast spo11Δ (null) mutants lack chromosome pairing, but catalytic site tyrosine mutants (spo11-Y135F) support significant levels of DSB-independent pairing [29], so the function of Spo11 proteins in pairing may be separable from its function in catalyzing recombination. Localization of Spo11 across synapsed, pachytene chromosomes of mice also suggests that Spo11 has an additional role, after it catalyzes recombination, in meiosis [25].
This study examined the relationship between meiotic recombination and chromosome segregation in rec12 (spo11) mutants of fission yeast. We report that Rec12 protein and its active site tyrosine are essential for crossover recombination in multiple intervals of all three chromosomes. Rec12 also has two roles in reductional chromosome segregation during MI: First, catalytically-active Rec12 is required for normal levels of reductional (chiasmatic) segregation. Second, catalytically-inactive Rec12 protein facilitates achiasmatic (distributive) segregation. In addition, we report that Rec12 and its active site tyrosine are required for proper equational chromosome segregation during MII.
Results
Structure of rec12+ gene and complementation by rec12+ cDNA
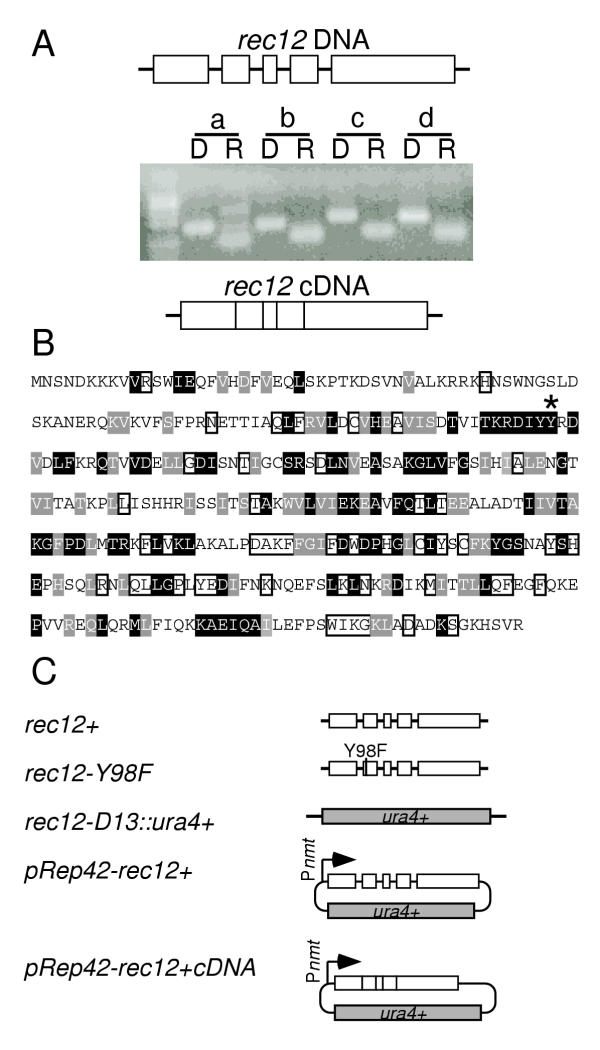
While rec12+ was reported to encode a protein of 139 amio acids [30], conceptual translation of the surrounding region suggested that rec12+ encoded a larger protein with homology to Spo11 of S. cerevisiae and Top6A of Sulfolobus shibatae [17,21]. Alignments and splicing sequence profiles [31] were used to identify four putative introns (Figure 1A). Oligonucleotide primer pairs flanking each of the putative introns were used for PCR and RT-PCR to amplify genomic DNA and meiotic mRNA, respectively. In each case, the product from mRNA was shorter than that from genomic DNA, confirming the presence of four introns (Figure 1A).
Figure 1.
Structure of the rec12 gene and constructs used. (A) Intron/exon structure. Genomic DNA (D) and total RNA (R) obtained from meiotic cultures of strain WSP0020 were subject to PCR and RT-PCR, respectively, using primers flanking the putative introns (a-d). Products were resolved on a 2% agarose gel stained with EtBr. (B) Primary sequence of Rec12 protein based upon the DNA sequence of a complementing cDNA clone. Residues of Rec12 with at least 50% identity (black boxes) or 50% conservation (gray boxes) relative to other eukaryotic Spo11 family members are indicated. Also shown are the positions of the active site tyrosine (*) and residues conserved among other Spo11 family members, but not conserved in S. pombe (open boxes). Alignments evaluated proteins from S. pombe (P40384), Neurospora crassa (Q9P6Y7), Coprinus cinereus (Q9P4D2), Homo sapiens (Q9NQM7), Mus musculus (Q9QZS1), Arabidopsis thaliana (AAL01152), Drosophila melanogaster (O77205), Caenorhabditis elegans (Q22236), and Saccharomyces cerevisiae (P23179). (C) Structure of constructs. Gene targeting of the endogenous rec12+ locus was used to introduce a null allele (rec12-D15::ura4+) lacking the complete coding region and a point mutation allele (rec12-Y98F) encoding a protein in which the active site tyrosine at position 98 was replaced with phenylalanine. Placing the rec12+ coding region and the rec12+ cDNA into the various pREP plasmids [32] allowed for a wide range of regulated gene expression.
Presence of four introns suggested that the rec12+ cDNA encodes a protein of 345 amino acids with high homology to eukaryotic Spo11 proteins (Figure 1B). To test this hypothesis, we introduced a full-length genomic clone and a sequence-confirmed cDNA into inducible expression vectors [32] in such a way that the first methionine in predicted exon 1 would be used for translation (Figure 1C). Each of these constructs restored wild-type levels of recombination to rec12-117 strains (Figure 2A, 2B). We conclude that the cDNA encodes a functional Rec12 protein of 345 amino acids in length. This conclusion was confirmed by Western blotting of an epitope-tagged Rec12 protein expressed from the endogenous rec12+ locus: a meiotically-induced protein band of the expected size was observed (W.D. Sharif and W.P. Wahls, unpublished observations).
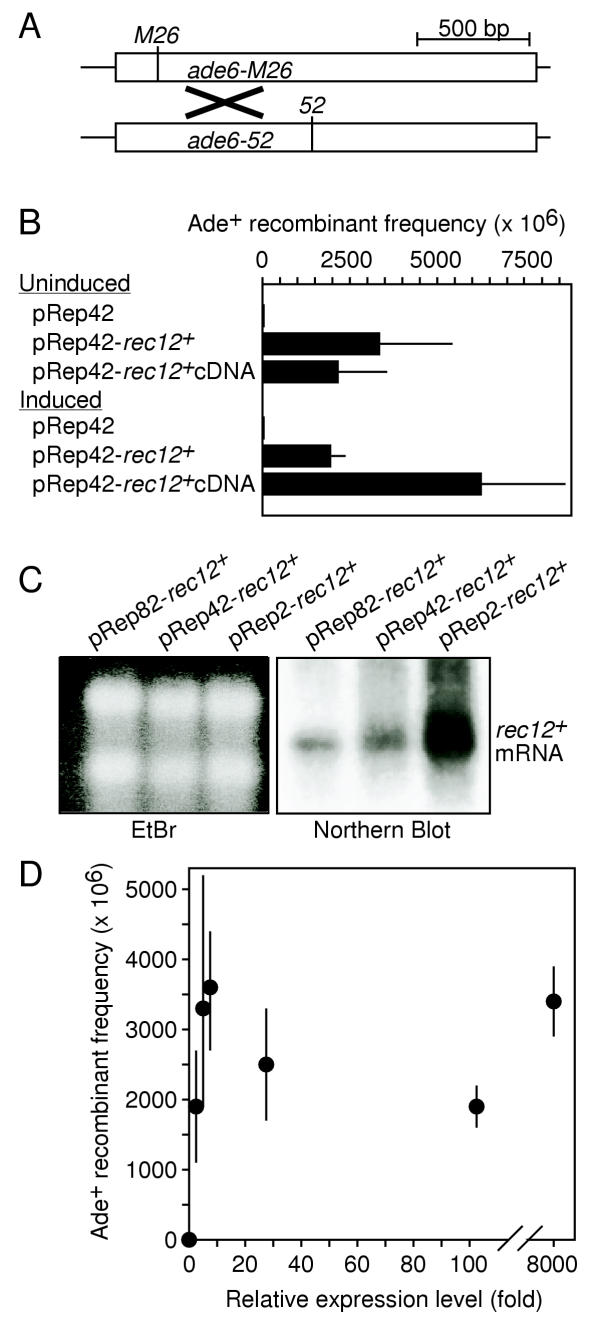
Figure 2.
Complementation of hyporecombination phenotype of rec12-117 mutants by rec12+ cDNA and overexpression of rec12+ gene. (A) Recombination substrates. Intragenic recombination between the ade6-M26 and ade6-52 alleles was measured. (B) Complementation by rec12+ and full-length rec12+ cDNA. Assays were with rec12-117 strains harboring pREP42 inducible expression vector [32] constructs. Data are mean ± standard deviation from three separate experiments involving crosses of strains WSP1065 × WSP1067; WSP1066 × WSP1068; and WSP1823 × WSP1824. (C) Northern blot analysis of rec12+ gene expression induced from strains harboring low-, middle-, and high-expression versions of pREP vectors. Data were obtained using strains WSP1058; WSP1066; and WSP1074. (D) Effect of rec12+ expression level upon recombination in rec12-117 strain. Recombinant frequency data are the mean ± standard deviation from three separate experiments; expression levels are β-galactosidase levels of similar constructs under non-inducing and inducing conditions [32]. Relative rec12+ expression levels were: pREP2u (u = uninduced; i = induced) < pREP82-rec12+,u < pREP42-rec12+,u < pREP82-rec12+,i < pREP42-rec12+,i < pREP2-rec12+,u < pREP2-rec12+,i. Data were obtained from crosses of strains WSP1073 × WSP1075; WSP1058 × WSP1060; WSP1066 × WSP1068; and WSP1074 × WSP1076.
Rec12 (Spo11) protein shares homology with the catalytic (Top6A) subunit of Topoisomerase VI, which functions as a heterotetramer with Top6B [16,17]. However, no Top6B homolog is present in most eukaryotes and the Rec12 (Spo11) enzyme does not need to affect strand passage in the manner of canonical type II topoisomerases [11]. We therefore tested whether overexpression of rec12+ would lead to a hyper-recombination phenotype, thereby implicating function as a homodimer. Very low levels of ectopic rec12+ expression restored wild-type recombination levels to the rec12-117 mutant (Figure 2B), confirming that functional Rec12 protein was produced. However, upon overexpression of rec12+ the frequency of recombinants did not increase significantly above wild-type levels (Figure 2C, 2D). Thus, Rec12 protein in excess of wild-type levels is biologically inactive; some other factor becomes rate-limiting under these circumstances.
The rec12-117 mutant was isolated in a genetic screen; the rec12-117 mutation reportedly confers a 100-fold or greater reduction of intragenic recombination and a 6-fold reduction of intergenic recombination [33]. To define more precisely the requirement for Rec12 in meiotic chromosome dynamics, we used gene replacement to generate a null (rec12-D15) mutation and a mutation in the putative active site tyrosine (rec12-Y98F). We then determined the effects of these mutations upon intergenic (crossover) recombination within intervals of chromosomes I, II, and III. We also employed a combination of cytological and genetic approaches to reveal the nature of meiotic chromosome segregation defects for each chromosome in each of the three mutants.
Rec12 and its active site tyrosine are required for chiasmatic meioses
Models for recombination posit that gene conversion and reciprocal exchange (crossover recombination) are alternative outcomes of a common intermediate [11]. Intragenic recombination at the ade6 locus, a measure of gene conversion, was reduced more than 100-fold in rec12-117 mutants [33], and we have obtained similar results with the rec12-117, rec12-D15, and rec12-Y98F mutants (Figure 2; W.D. Sharif and W.P. Wahls, unpublished data). Because reciprocal exchange accompanies more than half of all gene conversions at the ade6 locus [34,35], the rec12 mutants must also have a reduced number of reciprocal exchanges in the vicinity of ade6.
If Rec12 protein is required for all (or most) meiotic recombination, then rec12 mutants might fail to produce a sufficient number of reciprocal exchanges to ensure that each chromosome pair receives at least one crossover. To address this issue, we determined the number of crossovers present in regions of chromosomes I, II, and III in the rec12-D15 (deletion) and rec12-Y98F (active site) mutants. Multiple genetic intervals encompassing approximately 20% of the genome (444 cM) were analyzed.
Intergenic recombination was reduced an average of 256-fold in the rec12-D15 (null) mutants and 263-fold in the rec12-Y98F (active site) mutants, relative to wild-type levels (Table 1). Thus, Rec12 protein and its active site tyrosine are likely required for most crossover recombination throughout the genome. Because there are between 11 and 18 crossovers per chromosome in wild-type S. pombe meioses, and those crossovers are distributed without interference [36], we can estimate the residual number of crossovers present on each chromosome in the rec12 mutant meioses. A 250-fold reduction in recombination would leave less than 0.1 crossovers per chromosome, so all three chromosomes in most of the rec12 mutant meioses are achiasmatic.
Table 1.
Intergenic (crossover) recombination in rec12 mutants.
| Genetic interval (chromosome) | No. testeda | No. recombinanta | Recombinant freq. (%) | cMb | cM in wild-typec | Fold reductionc |
| A. rec12-D15 | ||||||
| lys1 – his1 (I) | 500 | 3 | 0.6 | 0.6 | 89 | 148 |
| lys1 – ade4 (I) | 500 | 8 | 1.6 | 1.6 | 284 | 177 |
| his1 – ade2 (I) | 500 | 2 | 0.4 | 0.4 | 57 | 142 |
| his1 – tps19 (I) | 500 | 2 | 0.4 | 0.4 | 143 | 357 |
| ade2 – tps19 (I) | 500 | 0 | ≤ 0.2 | ≤ 0.2 | 86 | ≤ 430 |
| his4 – ade1 (II) | 500 | 6 | 1.2 | 1.2 | 63 | 53 |
| ade6 – arg1 (III) | 500 | 1 | 0.2 | 0.2 | 97 | 486 |
| mean = | 256 ± 154 | |||||
| B. rec12-Y98F | ||||||
| lys1 – his1 (I) | 500 | 3 | 0.6 | 0.6 | 89 | 148 |
| lys1 – ade4 (I) | 500 | 17 | 3.4 | 3.5 | 284 | 81 |
| his1 – ade2 (I) | 500 | 1 | 0.2 | 0.2 | 57 | 285 |
| his1 – tps19 (I) | 500 | 1 | 0.2 | 0.2 | 143 | 715 |
| ade2 – tps19 (I) | 500 | 0 | ≤ 0.2 | ≤ 0.2 | 86 | ≤ 430 |
| his4 – ade1 (II) | 500 | 5 | 1.0 | 1.0 | 63 | 63 |
| ade6 – arg1 (III) | 500 | 4 | 0.8 | 0.8 | 97 | 121 |
| mean = | 263 ± 220 | |||||
aSpore colonies were replica plated on non-selective and selective media; the number of colonies recombinant for each genetic interval are indicated. Diploid (potentially complementing) spore colonies were excluded from analysis. bGenetic distance calculated using the mapping function of Haldane [66]. cFold reduction in crossing over relative to published map distances in wild-type cells (NCBI Entrez Genome) [36]. The strains crossed were: WSP1826 × WSP1967; WSP1825 × WSP1962; WSP1967 × WSP1973; WSP1967 × WSP1973; WSP1967 × WSP1973; WSP2015 × WSP2018; WSP2026 × WSP2024; WSP2020 × WSP1966; WSP1670 × WSP1968; WSP1966 × WSP2027; WSP1966 × WSP2027; WSP1966 × WSP2027; WSP2014 × WSP2011; WSP1809 × WSP2028. Strain genotypes are provided in Table 3.
Rec12 is required for proper meiotic chromosome segregation
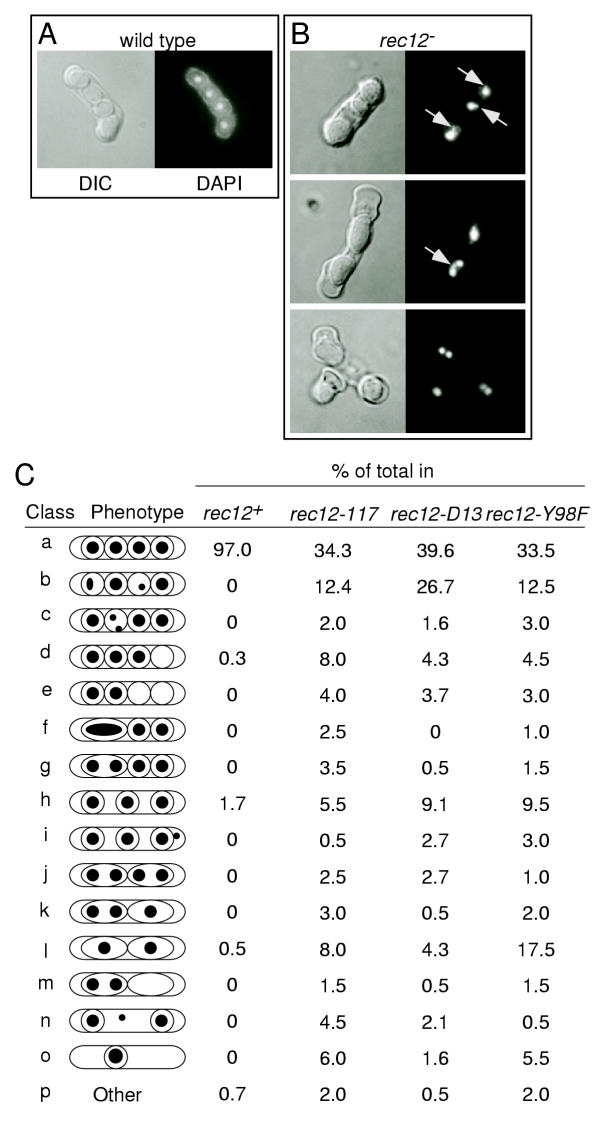
The rec12 mutant meioses are achiasmatic and would thus be expected to suffer chromosome segregation errors during meiosis I. To gain insight into this possibility, asci from meiotic cultures were stained with a DNA specific fluorescent dye (DAPI) and visualized by differential interference contrast (DIC) and fluorescence microscopy. The majority of asci from wild-type cells contained four well-rounded spores, each with a single DAPI-staining body of equal intensity (Figure 3A). The rec12 mutants were proficient at meiosis and underwent two meiotic divisions as revealed by ascus development and the distribution of chromosomes. However, each of the rec12 mutants exhibited a high frequency of chromosome segregation errors that were sometimes accompanied by defects in spore formation and/or ascus development (Figure 3B). The data from a large number of asci from each mutant are presented schematically in Figure 3C.
Figure 3.
Cytological phenotypes of wild-type and rec12 mutant asci. (A-B) DIC (left) and DAPI fluorescence (right) images of asci. In addition to aberrant ascus morphology, variable spore number, and unequal DNA content, some rec12 mutant asci show evidence of trailing DNA and chromatin bridges (arrows). The data were from crosses of strains: WSP0602 × WSP0603; WSP0332 × WSP0335 or WSP0079 × WSP1799; WSP1813 × WSP1819; and WSP1556 × WSP1559. (C) Summary of cytological phenotypes in wild-type, rec12-117, rec12-D15, and rec12-Y98F. Randomly selected asci were classified based upon DNA content and distribution (black dots), spore coat formation (circles), and ascus morphology (peripheral oval) using data such as in Panel B. Between 187 and 300 asci were scored for each group. Photomicrographs corresponding to classes "f," "l," and "j" are shown in Panel B.
The rec12-117, rec12-D15, and rec12-Y98F mutants exhibited qualitatively and quantitatively similar cytological phenotypes (Figure 3C). They each produced between 60% and 66% of asci with gross defects in DNA distribution. These mutant phenotypes were not uniform; different asci from the same cross were differentially affected (Figure 3). This heterogeneous phenotype is characteristic of a variety of mutations that directly affect meiotic chromosome dynamics [37-39]. Aberrant (or random) assortment of the three chromosome pairs (or twelve chromatids) leads to heterogeneous DNA distributions within different asci.
The rec12 mutants also exhibited high frequencies of additional developmental defects such as abnormal spore placement, reduced spore numbers, and abnormal spore formation (Figure 3C). It has been suggested previously [38] that these phenotypes are characteristic of a defect in meiosis II because spore formation in S. pombe is controlled by the spindle pole body of the MII spindle [40-42].
S. pombe produces ordered tetrads. If chromosomes segregated aberrantly during the MI reductional division, and then segregated appropriately during the equational division of MII, one would expect sister spores to receive identical complements of chromosomes. However, among rec12 mutant asci with four spores and an aberrant DNA distribution (Figure 3C, classes b, c, d, e), only 26% or less exhibited a pattern characteristic of simple MI nondisjunction (Figure 3C, classes c, e). The remaining 74% or more had DNA distributions characteristic of precocious sister chromatid separation and/or MII segregation errors – DNA was unequally distributed between two sister spores (Figure 3C, classes b, d). In some cases a defect in chromosome segregation during MII was visualized more directly. For example, in many cases DNA masses failed to separate properly into two sister spores during the second meiotic division (Figure 3B; Figure 3C, classes f, g, j, k, m). Thus, Rec12 protein is required for some function of MII, as well as for recombination during MI.
rec12 mutants produce a high frequency aneuploid meiotic products
Since S. pombe contains only three chromosome pairs, aberrant assortment during one or both of the meiotic divisions can produce, by chance, a significant fraction of meiotic products that receive at least one copy of each chromosome and are hence viable [38,43]. Aberrant segregation can also produce nullisomic aneuploids (missing one or more chromosomes), polysomic aneuploids (having additional copies of individual chromosomes), and diploids [38,39,44]. In S. pombe the haploids and diploids are viable, nullisomic aneuploids are inviable, and disomic aneuploids tend to lose the extra chromosome or chromosomes and become haploid [45]. We therefore determined the frequencies of spore viability and meiotic diploidy as genetic measures of chromosome segregation errors.
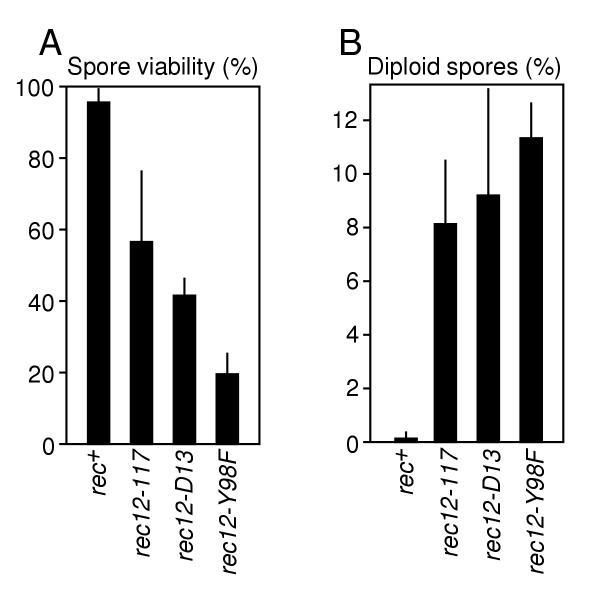
Approximately 50% of the meiotic products from the rec12-117 and rec12-D15 (null) mutants were inviable (Figure 4A), indicating that about 50% of the products were nullisomic for one or more chromosomes. This value is close to the frequency (58%) of nullisomics that one would expect if chromosome segregation were completely random in either one of the two meiotic divisions. Interestingly, the rec12-Y98F mutant produced a significantly higher frequency of inviable spores than the rec12-117 and rec12-D15 mutants (Figure 4A), suggesting that rec12-Y98F is a separation of function mutation.
Figure 4.
Formation of aneuploid meiotic products. (A) Spore viabilities. (B) Frequencies of diploid meiotic products. Data are the mean ± standard deviation from six separate experiments involving crosses of strains WSP0602 × WSP0603; WSP0079 × WSP1799; WSP1813 × WSP1819; and WSP1556 × WSP1559.
As a second measure of aberrant segregation, we determined the frequency of diploid spore colonies in the mutants. Each of the rec12 mutants behaved similarly and produced approximately 10% diploid meiotic products, more than 30-fold higher than the frequency produced by wild-type cells (Figure 4B). If chromosome segregation were completely random in either one of the two meiotic divisions, one would expect approximately 4% of the viable products to be disomic for all three chromosomes. The value obtained in the rec12 mutants was 2.5-fold higher than the predicted random value, indicating that the segregation errors are not simply the consequence of random chromosome assortment (nondisjunction) occurring exclusively during one of the two meiotic divisions.
Rec12 is required for chiasmatic and achiasmatic chromosome segregation during MI; and its active site tyrosine is required for chromosome segregation during MII
Because each of the rec12 mutants had aberrant meiotic chromosome segregation (Figure 3), produced a high frequency of viable meiotic products (Figure 4A), and produced a high frequency of meiotic diploids (Figure 4B), we were able to monitor directly the segregation patterns of all three chromosome pairs in each of the mutants. Meiotic diploids were genotyped for heteroallelic, centromere-linked markers on each of the three chromosomes. Those data were compared to the inheritance patterns predicted for each type of chromosome missegregation (Figure 5).
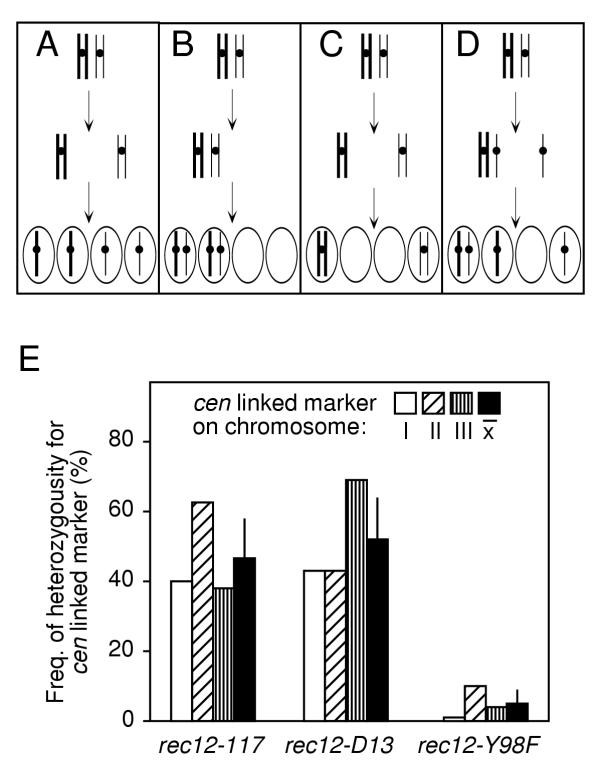
Figure 5.
Segregation patterns of chromosomes. (A) Normal meiosis produces four haploid products. Aberrant segregation may be monitored using centromere linked markers. Diploid spores resulting from MI nondisjunction (B) and precocious separation of sister chromatids (D) are predominantly heterozygous for the centromere, whereas those arising by MII nondisjunction (C) are homozygous for the centromere. (E) Diploid spore colonies were genotyped for heteroallelic marker loci tightly linked to each of the three centromeres (lys1, chromosome I; tps13, chromosome II; and ade6, chromosome III). Each value is based upon genotyping between 50 and 80 diploid spore colonies and the mean ± standard deviation for all three chromosomes in each mutant is indicated (black bars). The data were from crosses of strains: WSP1807 × WSP1216; WSP1825 × WSP1884; and WSP1670 × WSP1809.
Approximately half of the diploid products from the rec12-117 and rec12-D15 (null) meioses were heterozygous for centromere-linked markers on chromosomes I, II, and III (Figure 5E). Thus, those meioses did not exhibit simple MI nondisjunction, because 100% of diploids arising from MI nondisjunction would be heterozygous at the centromeres (Figure 5B). Precocious separation of sister chromatids during MI [44,46] can also produce diploid meiotic products, 67% of which would be heterozygous for centromere markers (Figure 5D) [38]. However, the average heterozygosities in the rec12-117 and rec12-D15 mutants were significantly below this value, indicating that these mutants must have suffered some proportion of MII segregation errors. By applying mapping functions for aneuploid meiotic products [47], we can set a lower limit of 22% and 29% MII nondisjunction for the rec12-117 and rec12-D15 meioses analyzed, respectively.
Surprisingly, the rec12-Y98F (active site) mutants produced meiotic diploids that were almost exclusively homozygous for centromere-linked markers (Figure 5E). The average heterozygosity for loci on all three chromosomes was 4.7 ± 4.1%. This indicates that about 95% of those rec12-Y98F mutant cells that produced diploid spores must have successfully completed the reductional segregation of chromosomes (MI) and failed to properly segregate sister chromatids during the equational division (MII).
While there was clear cytological evidence of two meiotic divisions and aberrant chromosome assortment in each division in the majority of rec12 mutant cells (Figure 3), it remained possible that some of the diploid meiotic products were derived from cells that had skipped one of the two meiotic divisions, thereby biasing the chromosome segregation data. If this were the case, all three chromosomes within any given, individual meiotic product would exhibit identical segregation patterns. However, about half of the diploids from rec12-117 and rec12-D15 meioses, and 14% of diploids from rec12-Y98F meioses, exhibited mixed patterns of MI or MII missegregation for different chromosomes within individual cells (Table 2). Furthermore, the observed frequencies of mixed segregation matched the frequencies predicted from the average values for single chromosomes (e.g., 14% vs. 13% for rec12-Y98F). These data provide additional evidence that two meiotic divisions had occurred and that the diploids arose as a consequence of meiotic chromosome segregation defects.
Table 2.
Aberrant segregation patterns of all three chromosomes within individual meiotic products.
| Type of segregation errora | Number of spores with segregation error | ||||
| Chr. I | Chr. II | Chr. III | rec12-117 | rec12-D15 | rec12-Y98F |
| MI | MI | MI | 12 | 18 | 0 |
| MII | MI | MI | 9 | 4 | 0 |
| MI | MII | MI | 0 | 3 | 0 |
| MII | MII | MI | 9 | 24 | 1 |
| MI | MI | MII | 10 | 5 | 0 |
| MII | MI | MII | 14 | 1 | 6 |
| MI | MII | MII | 7 | 3 | 0 |
| MII | MII | MII | 19 | 9 | 43 |
| n= | 80 | 67 | 50 | ||
aThe inferred type of segregation error for each chromosome within each diploid spore colony was determined by the analysis of centromere-linked markers as in Figure 5E. MI, heterozygous; MII, homozygous centromere-linked marker. The strains crossed were: WSP1807 × WSP1216; WSP1825 × WSP1884; and WSP1670 × WSP1809. Strain genotypes are provided in Table 3.
Discussion
Biochemical and genetic approaches identified the active site tyrosine of budding yeast Spo11 protein [17,21]. We introduced an active site mutation (rec12-Y98F), as well as a null mutation (rec12-D15), into the rec12 gene of S. pombe; fission yeast is the second organism in which studies of cells expressing catalytically-inactive Spo11 (Rec12) have been reported. This, coupled with the biology of fission yeast, enabled us to explore functions of Rec12 in a way that is not readily possible in other organisms.
There are two key features of fission yeast biology that allowed us to identify separate functions of Rec12 protein and Rec12-dependent crossovers in meiotic chromosome segregation. First, none of the many meiotic recombination deficient mutants of fission yeast identified so far confer a meiotic arrest phenotype [48]. The rec12 mutant cells complete meiosis and the majority show clear evidence of two meiotic divisions (Figure 3, Table 2), thus allowing us to study chromosome segregation in both divisions. This is in contrast to spo11 mutants of other organisms, such as mice and Coprinus cinereus, in which apoptosis occurs at or after meiotic prophase [24,25,49]. Second, for mutations affecting chromosome segregation the probability of obtaining viable meiotic products decreases with the power of chromosome number. As a consequence, spo11 (rec12) mutants of organisms with many chromosomes (e.g., budding yeast) produce almost exclusively inviable meiotic products [50]. In contrast, S. pombe contains only three chromosomes, so if chromosome segregation were entirely random in either one of the two meiotic divisions, 42% of the meiotic products would receive one or more copies of each of the three chromosomes and hence be viable {P = (0.75)3}. These two features of fission yeast allowed us to monitor directly the effects of different rec12 alleles upon recombination and chromosome segregation.
Conserved structure and function of Rec12 (Spo11) to initiate recombination
Identification of four introns (Figure 1) and complete complementation of the rec12-117 hyporecombination defect by the full-length cDNA (Figure 2) confirmed that Rec12 protein is homologous to Spo11 of other eukaryotes [11]. Both Rec12 and its active site tyrosine are required for intragenic and intergenic recombination at multiple locations on all three chromosomes (Figure 2, Tables 1, 2, data not shown). We observed an average, 256-fold and 263-fold reduction in crossover recombination in the rec12-D15 and rec12-Y98F mutants for intervals encompassing ~20% of the genome. Since wild-type S. pombe has 44 crossovers per meiosis (between 11 and 18 per chromosome) [36], the mutants statistically have less than 0.1 crossovers per chromosome per meiosis and are thus achiasmatic.
It was reported previously that rec12-117 mutants exhibit only a 6-fold reduction in crossover recombination [33], which is at odds with our value of more than a 250-fold reduction in the rec12-D15 (null) and rec12-Y98F (active site) mutants (Table 1). This discrepancy is not due to the nature of the rec12-117 allele, as we have also observed a 200-fold or greater reduction in crossovers in rec12-117 mutants (W.D. Sharif and W.P. Wahls, unpublished data). Our observation that rec12-117 mutants produce a high frequency of viable diploid meiotic products, about half of which are heterozygous for centromere linked markers (Figures 4, 5, Table 2), provides a straightforward reason for the discrepancy: The previous study failed to exclude diploid spore colonies and scored only for double prototrophic recombinants (the double auxotrophic, reciprocal recombinants were not analyzed). Presumably the bulk of the colonies reported previously as "recombinants" were actually diploids (or disomics) exhibiting intergenic complementation.
We used dosage studies to see if Rec12 functions in a manner predicted by homology to Spo11/Top6A. Upon overexpression of Rec12 protein, the frequency of recombination did not increase beyond wild-type levels (Figure 2). The simplest interpretation is that Rec12 functions in a multisubunit protein complex; a complex in which other components are required either for efficient catalysis of dsDNA breaks or for correct subcellular localization of Rec12. In budding yeast the active site mutation exhibits allele-specific semidominance when heterozygous with wild-type [51], and we have made similar observations in fission yeast (W.D. Sharif and W.P. Wahls, unpublished data). This provides evidence that a dimer of Rec12/Spo11 protein, harboring two functional active sites, is required for catalysis of dsDNA breaks in each organism. Thus, with regard to primary protein sequence (Figure 1), inferred subunit structure (Figure 2 and data not shown), strict requirement for meiotic recombination (Figure 2, Table 1), and dependence upon its active site tyrosine (Table 1), Rec12 protein is a structurally and functionally conserved member of the Spo11 family of enzymes that catalyze meiotic dsDNA cleavage to initiate recombination [11].
Rec12 has dual roles in chiasmatic and achiasmatic (distributive) chromosome segregation during Meiosis I
In the absence of crossover recombination (chiasmata) and a backup distributive (achiasmatic) system for chromosome segregation, MI nondisjunction should occur and 100% of any resulting diploid spore colonies would be heterozygous for centromere linked markers (Figure 5B). However, only about 50% of diploid meiotic products from rec12-D15 (null) mutant meioses were heterozygous for centromere linked markers (Figure 5E), indicating that some fraction of the products were derived from precocious separation of sister chromatids (PSS) and/or MII nondisjunction (Figure 5C, 5D). Since PSS does not occur in rec12 null mutants [52], those centromere-homozygous chromosomes must have experienced a proper reductional segregation and a failed equational segregation (discussed below).
The rec12-D15 (null) mutants are achiasmatic (Table 1) and, as anticipated, exhibit a high frequency of MI nondisjunction (Figures 3, 5, Table 2). However, the frequency of MI nondisjunction observed is only half of that expected, suggesting the presence of a distributive (achiasmatic) system of chromosome segregation. There is precedence for a distributive MI segregation system in several organisms. [7,53]. In rec7 hyporecombination mutants of S. pombe the fidelity of segregation of homologs during MI exceeds the expected random level [10]. Similarly, MI segregation of GFP-tagged centromeres in rec12 deletion mutants occurs at non-random levels (L. Davis and G.R. Smith, personal communication), in good agreement with our genetic results. In each case identified, this distributive system of fission yeast can partially circumvent the MI segregation errors of achiasmatic chromosomes; approximately one-quarter to one-half of those homologs expected to nondisjoin (i.e., move to the same pole) end up segregating distributively (i.e., move to opposite poles).
While the rec12-D15 (null) mutants frequently suffer MI nondisjunction, the rec12-Y98F (active site) mutants produce meiotic diploids with nearly normal MI segregation patterns (Figures 3, 5, Table 2). This difference is unrelated to crossover frequency, because the active site and null mutants have identical deficiencies in crossover recombination (Table 1). Thus, Rec12 protein has one or more functions in MI that are distinct from its functions to catalyze recombination and mediate chiasmatic segregation. We propose that the Rec12-Y98F protein facilitates distributive segregation of achiasmatic chromosomes during MI.
A meiotic checkpoint could provide additional time for missegregating chromosomes to correct their erroneous wanderings during MI [10]. We propose two likely mechanisms by which the Rec12 protein improves the MI segregation of achiasmatic chromosomes. First, Rec12 may be required for an "MI nondisjunction checkpoint" that delays meiotic cells in which proper MI disjunction has failed to occur. If Rec12 were required for such a checkpoint, the null mutants would be achiasmatic, would fail to delay for the time period required to establish distributive segregation, and would consequently undergo high rates of MI nondisjunction. In the presence of Rec12-Y98F protein the checkpoint would be active, thereby allowing the achiasmatic chromosomes sufficient time to reorient and undergo distributive segregation. Meiotic progression in spo11 (rec12) null mutants of budding yeast is accelerated [29], supporting our hypothesis that Rec12 may participate in an "MI nondisjunction checkpoint."
As an alternative (but not mutually exclusive) mechanism, Rec12 protein may participate directly in the machinery of distributive segregation. In female meioses of D. melanogaster, heterochromatic pairing of homologs can substitute for chiasmata to ensure bipolar spindle attachment of bivalents and subsequent distributive segregation [4-6,8]. Rec12 may serve a similar role in maintaining a paired (or pseudo-paired) state between achiasmatic chromosomes prior to the first meiotic division. In support of this possibility, Spo11 of mice colocalizes extensively along homologously synapsed chromosomes during pachytene and dissociates when the homologs segregate from one-another during anaphase of MI [25].
Rec12 has a role in equational chromosome segregation during Meiosis II that is dependent upon its active site tyrosine
Cytological and genetic data each demonstrated that the rec12-D15 (null) and rec12-Y98F (active site) mutants frequently suffer chromosome segregation errors during MII (Figures 3, 5, Table 2). This role for Rec12 in equational chromosome segregation may be conserved, as spo11 mutants of Sordaria macrospora also exhibit frequent MII nondisjunction (S. Tesse, A. Storlazzi, S. Gargano, and D. Zickler, personal communication). Irregular partitioning of chromosomes during the second meiotic division has also been reported in fission yeast rec7 mutants [10], so MII missegregation may be a relatively common fate for chromosomes in hyporecombination mutants. As described previously in this paper and elsewhere [38], the biology of fission yeast facilitates the identification of MII segregation defects that are difficult to detect in other organisms that terminate meiotic progression or produce inviable meiotic products as a consequence of defects in meiotic chromosome dynamics.
Although they exhibit a striking difference in their apparent MI nondisjunction frequencies, the rec12-D15 (null) and rec12-Y98F (active site) mutants manifest at most a two-fold difference in the absolute frequency of meiotic diploids exhibiting MII missegregation patterns (Figures 4, 5, Table 2). Catalytically-inactive Rec12-Y98F protein apparently cannot enhance the fidelity appropriate MII segregation, relative to MII missegregation levels in cells lacking Rec12 entirely. We therefore propose that Rec12 protein mediates a signal, strictly dependent upon its active site tyrosine, that is required for the equational division of MII.
There are two main hypotheses as to the nature of the signal: First, Rec12 protein itself may participate in the signal transduction. In pat1-114ts meioses Rec12 appears to persist until about the time of the second meiotic division (N. Herrera and S. Forsburg, personal communication), so Rec12 is available at the right time. Alternatively, some aspect of the crossovers or chiasmata may transduce the signal. This could involve structural components that persist on chromosomes at the positions where recombination and chiasmata had previously occurred, or could involve a diffusible component generated by recombination events.
At present, two hypothetical mechanisms by which a Rec12-dependent signal affects MII are proposed: First, Rec12-dependent (recombination-dependent) signals might trigger a block after MI, thereby allowing cells to skip a frequently irregular MII division [39]. However, the majority of the rec12 mutant meioses show evidence of two meiotic divisions (Figure 3) and many of the meiotic diploids exhibit mixed MI and MII missegregation patterns for centromere linked markers on the three different chromosomes (Table 2). In the absence of PSS, which does not occur at significant levels in rec12 mutants [52], such mixed segregation patterns can only be generated by a combination of errors in both reductional and equational segregation. A second hypothetical mechanism takes into account the coordinate, two-step release of sister chromatid cohesion that is essential for the two meiotic divisions [38]. Rec12-dependent (or recombination-dependent) signals may be required for the proper maintenance and/or dissolution of centromere-proximal sister chromatid cohesion in MII. These and other hypotheses regarding the unambiguous requirement for Rec12 in MII remain to be tested.
Conclusions
The role of Spo11 (Rec12) in the initiation of meiotic recombination via catalysis of dsDNA breaks is conserved and likely universal among eukaryotes. The rec12 null and active site mutants are achiasmatic and, as anticipated, Rec12-dependent crossovers (chiasmata) are required to ensure proper reductional chromosome segregation in MI. In the absence of Rec12 protein and crossovers, a backup distributive (achiasmatic) segregation system operates successfully on up to half of the chromosomes that would otherwise undergo MI nondisjunction. Presence of catalytically inactive Rec12 protein enhances the fidelity of this distributive segregation system. In addition, catalytically active Rec12 protein (or Rec12-dependent recombination) is required for faithful chromosome segregation during MII.
Methods
S. pombe culture
Strain genotypes are listed in Table 3. Culture media and genetic methods were as described [38,54,55].
Table 3.
Genotypes of S. pombe strains used for this study.
| Strain | Genotype |
| WSP 0020 | h+ade6-M26 pat1-114ts |
| WSP 0079 | h+ade6-M26 rec12-117 |
| WSP 0332 | h+ade6-M26 ura4-294 arg3-124 rec12-117 |
| WSP 0335 | h-ade6-52 ura4-595 pro2-1 rec12-117 |
| WSP 0602 | h+ade6-M26 ura4-D18 |
| WSP 0603 | h-ade6-M26 ura4-D18 |
| WSP 1058 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP82-rec12+) |
| WSP 1060 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP82-rec12+) |
| WSP 1065 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP42) |
| WSP 1066 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP42-rec12+) |
| WSP 1067 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP42) |
| WSP 1068 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP42-rec12+) |
| WSP 1073 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP2) |
| WSP 1074 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP2-rec12+) |
| WSP 1075 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP2) |
| WSP 1076 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP2-rec12+) |
| WSP 1216 | h-ade6-52 tps13-24 rec12-117 |
| WSP 1556 | h+ade6-M26 ura4-D18 his3-D1 leu1-32 rec12-Y98F |
| WSP 1559 | h-ade6-M26 ura4-D18 leu1-32 his3-D1 rec12-Y98F |
| WSP 1670 | h+lys1-131 rec12-Y98F |
| WSP 1799 | h-ade6-M26 rec12-117 |
| WSP 1807 | h+lys1-131 rec12-117 |
| WSP 1809 | h-ade6-52 rec12-Y98F tps13-24 |
| WSP 1813 | h-ade6-M26 ura4-D18 rec12-D15::ura4 |
| WSP 1819 | h+ade6-M26 ura4-D18 rec12-D15::ura4 |
| WSP 1823 | h+ade6-M26 ura4-294 arg3-124 rec12-117 (pREP42-rec12+cDNA) |
| WSP 1824 | h-ade6-52 ura4-595 pro2-1 rec12-117 (pREP42-rec12+cDNA) |
| WSP 1825 | h+lys1-131 rec12-D15::ura4 |
| WSP 1826 | h-lys1-131 rec12-D15::ura4 ura4-D18 |
| WSP 1884 | h-ade6-52 tps13-24 ura4-D18 rec12-D15::ura4 |
| WSP 1962 | h-ade4-31 rec12-D15::ura4 ura4-D18 |
| WSP 1966 | h+his1-102 rec12-Y98F |
| WSP 1967 | h+his1-102 ura4-D18 rec12-D15::ura4 |
| WSP 1968 | h-ade4-31 ura4-D18 rec12-Y98F |
| WSP 1973 | h-tps19-17 ade2-17 ura4-D18 rec12-D15::ura4 |
| WSP 2011 | h+ade1-51 rec12-Y98F |
| WSP 2014 | h-his4-239 rec12-Y98F |
| WSP 2015 | h-his4-239 ura4-D18 rec12-D15::ura4 |
| WSP 2018 | h+ade1-51 ura4-D18 rec12-D15::ura4 |
| WSP 2020 | h-lys1-131 rec12-Y98F |
| WSP 2024 | h-arg1-14 ura4-D18 rec12-D15::ura4 |
| WSP 2026 | h+ade6-52 rec12-D15::ura4 ura4-D18 |
| WSP 2027 | h-ade2-17 tps19-17 rec12-Y98F |
| WSP 2028 | h+arg1-14 rec12-Y98F |
Intron mapping, cDNA cloning, and DNA sequence analysis
Cells harboring the pat1-114ts allele were induced to enter meiosis [56] and were collected after 3 hours of meiotic induction. Genomic DNA and total RNA were prepared [57] and subject to PCR and RT-PCR [58] using primers flanking each putative intron.
Full-length rec12+ cDNA was obtained using RT-PCR. The 5' and 3' primers contained NdeI and BamHI restriction sites, respectively, and were designed to amplify the rec12+ cDNA from the first ATG in exon 1 to a position +142 bp downstream of the stop codon in exon 5. RT-PCR products were cloned by blunt-end ligation into pCR-Blunt (Invitrogen Corp.) and both strands were subject to DNA sequencing (GenBank accession no. AF195027).
Alignment of Rec12 with other Spo11 family members
Protein sequences homologous to that of S. pombe Rec12 were identified with a NCBI Blast search [59] using matrix BLOSUM62. Representative homologous sequences (see Figure 1 legend) were aligned using T-COFFEE [60], which optimizes alignment of sequences with low levels of homology. Output was prepared using a 50% threshold for identical and conserved residues.
Inducible pREP vector constructions
Genomic rec12+ DNA and rec12+ cDNA were cloned between the NdeI and BamHI restriction sites of the inducible vectors pREP2, pREP42, and pREP82 [32]. Induction of gene expression and northern blot analysis of rec12+ mRNA levels were as described [32,57].
Genomic DNA was liberated from pYL78 [30] by partial EcoRI digestion and complete BamHI digestion, which cleaves within the polylinker site approximately 700 bp downstream of the rec12+ stop codon in exon 5. The DNA fragment with single-site cleavage by EcoRI at bp position +3, relative to the ATG start codon, was gel purified. A dsDNA oligonucleotide linker, containing cohesive NdeI and EcoRI ends, was constructed (5'-TATGAGCCATCATCATCATCATCATAGCATG-3' and 5'-AATTCATGCTATGATGATGATGATGATGGCTAC-3'). This linker encoded in frame the amino acids MSHHHHHHSM, with the last residue corresponding to the first methionine encoded in rec12+ exon 1. Trimolecular ligation was used to create pREP2-rec12+, pREP42-rec12+, and pREP82-rec12+ clones encoding Rec12 protein with 6X histidine tags. An NdeI-BamHI fragment of a cDNA clone with wild-type rec12+ sequence (see above) was sub-cloned into the pREP42 vector to generate pREP42-rec12+cDNA. The integrity of all clones was determined by a combination of restriction mapping, PCR, and DNA sequencing.
Construction of rec12-D15::ura4+ and rec12-Y98F alleles
A PCR based gene targeting approach [61,62] was used to delete the rec12+ coding region and replace it with the ura4+ gene. Candidates were screened with a combination of PCR analysis, restriction digestion, and DNA sequencing to identify those with successful allele replacement.
Replacement of the endogenous rec12+ locus with the rec12-Y98F allele was achieved by transformation and a pop-in, pop-out approach [61,63]. A 1.2 kbp XbaI-XbaI fragment of rec12+ was subcloned into pBluescript SKII(+) and subject to oligonucleotide directed mutagenesis [64] to simultaneously introduce a silent mutation generating an EcoRV site and to mutate the Y98 codon (GAG ATA TTT ATT ACA → GAG ATA TCT ATT TCA). After confirmation by DNA sequencing, the XbaI-XbaI fragment was subcloned into pURA4. pURA4-rec12-Y98F was linearized by digestion with NcoI. Transformation, forward selection for Uracil prototrophy, and reverse selection for FOA resistance were as described [61,63]. Candidates were screened as described above.
Recombinant frequency determination
Mating, meiosis, and preparation of free spores were as described [54,55,65]. Intergenic and intragenic recombinant frequencies were determined as described [38,65]. Because diploid spores could contain complementing markers and be mistaken for recombinants, we tested all spore colonies for diploidy by replica plating on complete media containing Phloxin-B (YEA-B). On these plates, diploid cells produce dark pink colonies, whereas haploid cells produce light pink colonies [54]. Diploid meiotic products were excluded from recombinant frequency determinations.
Diploid spore isolation and haploidization analysis
Identification of diploid spore colonies and their haploidization with m-flourophenylalanine (m-FPA) [55] were as described [38]. Fifty haploidized colonies derived from each diploid spore colony were replica plated to differentially supplemented minimal media to genotype the lys1 and ade6 loci. The tps13 alleles were scored by replica plating colonies onto rich (YEA) media and testing for growth at permissive (22°C) and restrictive (35°C) temperatures.
Microscopy
Asci from meiotic cultures were fixed with 70% ethanol at -20°C for at least 15 minutes, washed with H2O, and stained with 4,6-diamidino-2-phenylindole (DAPI) at a final concentration of 1 μg/ml. Cells were examined by differential interference contrast (DIC) and fluorescence (DAPI) microscopy with a Zeiss axiophot (Carl Zeiss, Thornwood, NY). Images were analyzed using the MetaMorph software package (Universal Imaging, West Chester, PA).
Authors' contributions
Each author contributed significantly to the design and execution of the study; respective efforts are reflected by order of authorship.
Acknowledgments
Acknowledgements
We thank Jürg Kohli and Gerry Smith for providing strains; Susan Forsburg, Gerry Smith, and Denise Zickler (and their respective colleagues) for sharing unpublished results; Jeff Flick, Peter Kolodziej, Gisela Mosig, Neil Osheroff, and Harish Shandilya for helpful discussions; Michelle Krawchuk for initiating studies of cytology; and Jamie Rose and Shelia Gibbs for laboratory support. This work was supported by grants GM62244 from the National Institutes of Health and RG0075/1999-M from the Human Frontier Science Program. WDS was supported in part by a UNCF-Merck Graduate Science Research Dissertation Fellowship Award.
Contributor Information
Wallace D Sharif, Email: SharifWallaceD@uams.edu.
Gloria G Glick, Email: gloria.g.glick@vanderbilt.edu.
Mari K Davidson, Email: DavidsonMariK@uams.edu.
Wayne P Wahls, Email: WahlsWayneP@uams.edu.
References
- Hawley RS. Exchange and chromosomal segregation in eukaryotes. In: Kucherlapati R, Smith GR, editor. Genetic Recombination. American Society for Microbiology; 1988. pp. 497–527. [Google Scholar]
- Grell R. Distributive pairing. In: Ashburner M, Novitski E, editor. The Genetics and Biology of Drosophila. Academic Press; 1976. pp. 435–486. [Google Scholar]
- Carpenter AT. Distributive segregation: motors in the polar wind? Cell. 1991;64:885–890. doi: 10.1016/0092-8674(91)90313-n. [DOI] [PubMed] [Google Scholar]
- Hawley RS, Irick H, Zitron AE, Haddox DA, Lohe A, New C, Whitley MD, Arbel T, Jang J, McKim K, et al. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev Genet. 1992;13:440–467. doi: 10.1002/dvg.1020130608. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Hassold TJ. Human aneuploidy: lessons from achiasmate segregation in Drosophila melanogaster. Ann Hum Genet. 1998;62:467–479. doi: 10.1046/j.1469-1809.1998.6260467.x. [DOI] [PubMed] [Google Scholar]
- Carpenter AT. A meiotic mutant defective in distributive disjunction in Drosophila melanogaster. Genetics. 1973;73:393–428. doi: 10.1093/genetics/73.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson DS, Murray AW, Szostak JW. An alternative pathway for meiotic chromosome segregation in yeast. Science. 1986;234:713–717. doi: 10.1126/science.3535068. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Kohli J, Hiraoka Y. Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J Cell Sci. 2001;114:2843–2853. doi: 10.1242/jcs.114.15.2843. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/S1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Zenvirth D, Simchen G. Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr Genet. 2000;38:33–38. doi: 10.1007/s002940000126. [DOI] [PubMed] [Google Scholar]
- Bergerat A, Gadelle D, Forterre P. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J Biol Chem. 1994;269:27663–27669. [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implication for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. Covalent protein-DNA complexes at the 5' strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci USA. 1995;92:11274–11278. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- McKim KS, Hayashi-Hagihara A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 1998;12:2932–2942. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- Celerin M, Merino ST, Stone JE, Menzie AM, Zolan ME. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 2000;19:2739–2750. doi: 10.1093/emboj/19.11.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Smith GR. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ, Marr TG. Fission yeast gene structure and recognition. Nucleic Acids Res. 1994;22:1750–1759. doi: 10.1093/nar/22.9.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-D. [DOI] [PubMed] [Google Scholar]
- DeVeaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;136:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Krawchuk MD, DeVeaux LC, Wahls WP. Meiotic chromosome dynamics dependent upon the rec8+, rec10+, and rec11+ genes of the fission yeast Schizosaccharomyces pombe. Genetics. 1999;153:57–68. doi: 10.1093/genetics/153.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Hirata A. Ascospore development in the fission yeast Schizosaccharomyces pombe and S. japonicus. J Cell Sci. 1982;56:263–279. doi: 10.1242/jcs.56.1.263. [DOI] [PubMed] [Google Scholar]
- Shimoda C, Hirata A, Kishida M, Hashida T, Tanaka K. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1985;200:252–257. doi: 10.1007/BF00425432. [DOI] [PubMed] [Google Scholar]
- Hirata A, Shimoda C. Structural modification of spindle pole bodies during meiosis II is essential for normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast. 1994;10:173–183. doi: 10.1002/yea.320100205. [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Yanagida M. Triploid meiosis and aneuoploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;9:463–470. [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls WP. Centromere mapping functions for aneuploid meiotic products: analysis of rec8, rec10, and rec11 mutants of the fission yeast Schizosaccharomyces pombe. Genetics. 1999;153:49–55. doi: 10.1093/genetics/153.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox ME, Smith GR. Control of meiotic recombination in Schizosaccharomyces pombe. Prog Nucleic Acid Res Mol Biol. 1998;61:345–378. doi: 10.1016/s0079-6603(08)60831-4. [DOI] [PubMed] [Google Scholar]
- Merino ST, Cummings WJ, Acharya SN, Zolan ME. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc Natl Acad Sci USA. 2000;97:10477–10482. doi: 10.1073/pnas.190346097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S, Waddell CS, Esposito RE. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz RL, Alcid AD, Berger JM, Keeney S. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol Cell Biol. 2002;22:1106–1115. doi: 10.1128/MCB.22.4.1106-1115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Kakihara Y, Hiraoka Y, Nojima H. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 2001;20:3871–3881. doi: 10.1093/emboj/20.14.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KW. How meiotic cells deal with non-exchange chromosomes. Bioessays. 1994;16:107–114. doi: 10.1002/bies.950160207. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Smith GR. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- Kon N, Schroeder SC, Krawchuk MD, Wahls WP. Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hotspot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol Cell Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki ES. Amplification of RNA. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editor. PCR protocols: a guide to methods and applications. Academic Press, Inc.; 1990. pp. 21–27. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.3.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Francesconi S, Park H, Wang TS. Fission yeast with DNA polymerase delta temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in S. pombe. Proc Natl Acad Sci USA. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. The combination of linkage values, and the calculation of distances between loci of linked factors. J Genet. 1919;8:299–309. [Google Scholar]